7QID
 
 | | tentative model of the human insulin receptor ectodomain bound by three insulin | | Descriptor: | Insulin, Insulin receptor, Isoform Short of Insulin receptor | | Authors: | Gutman, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain
J Cell Biol, 219, 2020
|
|
6SOF
 
 | | human insulin receptor ectodomain bound by 4 insulin | | Descriptor: | Insulin, Insulin receptor | | Authors: | Gutmann, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
J.Cell Biol., 219, 2020
|
|
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CR2
 
 | | Homo sapiens Get1/Get2 heterotetramer (a3' deletion variant) in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8CQZ
 
 | | Homo sapiens Get3 in complex with the Get1 cytoplasmic domain | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor 1 | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Saar, D, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODV
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (nanodisc) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
8ODU
 
 | | Chaetomium thermophilum Get1/Get2 heterotetramer in complex with a Get3 dimer (amphipol) | | Descriptor: | ATPase GET3, Protein GET2,Protein GET1, ZINC ION | | Authors: | McDowell, M.A, Wild, K, Sinning, I. | | Deposit date: | 2023-03-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
6RSW
 
 | | HFD domain of mouse CAP1 bound to the pointed end of G-actin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Kotila, T, Kogan, K, Lappalainen, P. | | Deposit date: | 2019-05-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin.
Nat Commun, 10, 2019
|
|
6FM2
 
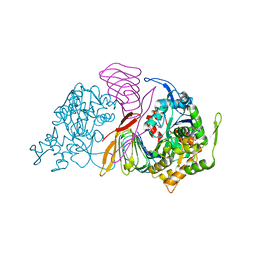 | | CARP domain of mouse cyclase-associated protein 1 (CAP1) bound to ADP-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kotila, T.M, Kogan, K, Lappalainen, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of actin monomer re-charging by cyclase-associated protein.
Nat Commun, 9, 2018
|
|
4D6A
 
 | |
4D6B
 
 | | Human myelin protein P2, mutant P38G | | Descriptor: | MYELIN P2 PROTEIN, PALMITIC ACID, SULFATE ION | | Authors: | Laulumaa, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2014-11-10 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Dynamics of the Peripheral Membrane Protein P2 from Human Myelin Measured by Neutron Scattering-A Comparison between Wild-Type Protein and a Hinge Mutant.
Plos One, 10, 2015
|
|
5N4Q
 
 | | Human myelin protein P2, mutant T51P | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4M
 
 | | Human myelin protein P2, mutant I43N | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4P
 
 | | Human myelin P2, mutant I52T | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
6EW4
 
 | |
6EW2
 
 | | Human myelin protein P2 F57A mutant, tetragonal crystal form | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Laulumaa, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Structure and dynamics of a human myelin protein P2 portal region mutant indicate opening of the beta barrel in fatty acid binding proteins.
BMC Struct. Biol., 18, 2018
|
|
6EW5
 
 | |
6RSQ
 
 | | Helical folded domain of mouse CAP1 | | Descriptor: | Adenylyl cyclase-associated protein 1, GLYCEROL | | Authors: | Kotila, T, Kogan, K, Lappalainen, P. | | Deposit date: | 2019-05-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin.
Nat Commun, 10, 2019
|
|
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
8C09
 
 | | Crystal structure of JAK2 JH2-I559F | | Descriptor: | Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
8C08
 
 | | Crystal structure of JAK2 JH2-K539L | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Haikarainen, T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
8C0A
 
 | | Crystal structure of JAK2 JH2-R683S | | Descriptor: | 3,5-diphenyl-2-(trifluoromethyl)-1~{H}-pyrazolo[1,5-a]pyrimidin-7-one, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Haikarainen, T, Silvennoinen, O. | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of JAK2 activation in erythropoietin receptor and pathogenic JAK2 signaling.
Sci Adv, 10, 2024
|
|
6XU5
 
 | | Human myelin protein P2 mutant N2D | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XUA
 
 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
