1CQW
 
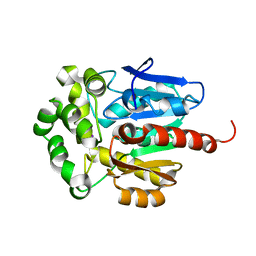 | | NAI COCRYSTALLISED WITH HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE; 1-CHLOROHEXANE HALIDOHYDROLASE, IODIDE ION | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1999-08-11 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1GKH
 
 | | MUTANT K69H OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1GVP
 
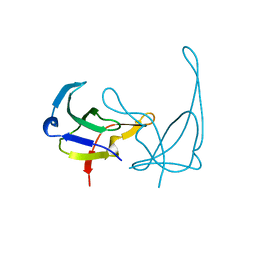 | | GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1AE2
 
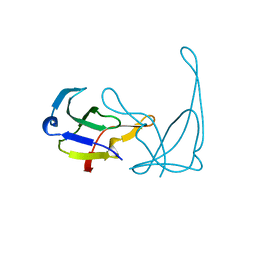 | | MUTANT L32R OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1AE3
 
 | | MUTANT R82C OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1BKB
 
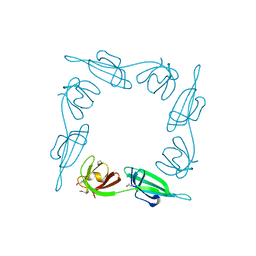 | | INITIATION FACTOR 5A FROM ARCHEBACTERIUM PYROBACULUM AEROPHILUM | | Descriptor: | TRANSLATION INITIATION FACTOR 5A | | Authors: | Peat, T.S, Newman, J, Waldo, G.S, Berendzen, J, Terwilliger, T.C. | | Deposit date: | 1998-07-05 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution.
Structure, 6, 1998
|
|
1BN7
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | ACETATE ION, HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BN6
 
 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
3TX8
 
 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6O7F
 
 | |
7TZ1
 
 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
1OY0
 
 | | The crystal Structure of the First Enzyme of Pantothenate Biosynthetic Pathway, Ketopantoate Hydroxymethyltransferase from Mycobacterium Tuberculosis Shows a Decameric Assembly and Terminal Helix-Swapping | | Descriptor: | Ketopantoate hydroxymethyltransferase, MAGNESIUM ION | | Authors: | Chaudhuri, B.N, Sawaya, M.R, Kim, C.Y, Waldo, G.S, Park, M.S, Terwilliger, T.C, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the First Enzyme in the Pantothenate Biosynthetic Pathway,
Ketopantoate Hydroxymethyltransferase, from M. tuberculosis
Structure, 11, 2003
|
|
1QP8
 
 | |
7WLS
 
 | | Crystal structure of the multidrug efflux transporter BpeB from Burkholderia pseudomallei | | Descriptor: | Efflux pump membrane transporter, TETRAETHYLENE GLYCOL, UNDECYL-MALTOSIDE | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WLV
 
 | | Crystal Structure of the Multidrug effulx transporter BpeF from Burkholderia pseudomallei. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8U22
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 7 | | Descriptor: | Green Fluorescent Protein Variant #7, ccGFP 7 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U21
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP E6 | | Descriptor: | Green Fluorescent Protein Variant E6, ccGFP E6 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U23
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 8 | | Descriptor: | Green Fluorescent Protein Variant #8, ccGFP 8 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
4FB7
 
 | | The apo form of idole-3-glycerol phosphate synthase (TrpC) form Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Indole-3-glycerol phosphate synthase | | Authors: | Michalska, K, Chhor, G, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The apo form of idole-3-glycerol phosphate synthase (TrpC) form Mycobacterium tuberculosis
To be Published
|
|
1VQH
 
 | |
1VQC
 
 | |
1VQF
 
 | | GENE V PROTEIN MUTANT WITH VAL 35 REPLACED BY ILE 35 AND ILE 47 REPLACED BY VAL 47 (V35I, I47V) | | Descriptor: | GENE V PROTEIN | | Authors: | Zhang, H, Skinner, M.M, Sandberg, W.S, Wang, A.H.-J, Terwilliger, T.C. | | Deposit date: | 1996-08-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context dependence of mutational effects in a protein: the crystal structures of the V35I, I47V and V35I/I47V gene V protein core mutants.
J.Mol.Biol., 259, 1996
|
|
1VQA
 
 | |
