4AGR
 
 | | Structure of a tetrameric galectin from Cinachyrella sp. (Ball sponge) | | Descriptor: | CHLORIDE ION, GALECTIN | | Authors: | Freymann, D.M, Focia, P.J, Sakai, R, Swanson, G.T. | | Deposit date: | 2012-01-31 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Tetrameric Galectin from Cinachyrella Sp. (Ball Sponge).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AGV
 
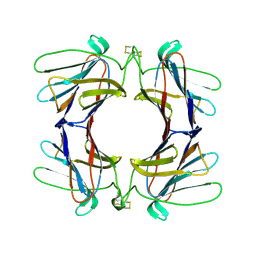 | | Structure of a tetrameric galectin from Cinachyrella sp. (Ball sponge) | | Descriptor: | GALECTIN | | Authors: | Freymann, D.M, Focia, P.J, Sakai, R, Swanson, G.T. | | Deposit date: | 2012-01-31 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Tetrameric Galectin from Cinachyrella Sp. (Ball Sponge).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AGG
 
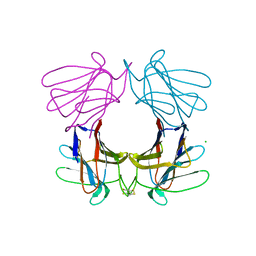 | | Structure of a tetrameric galectin from Cinachyrella sp. (Ball sponge) | | Descriptor: | CHLORIDE ION, GALECTIN | | Authors: | Freymann, D.M, Focia, P.J, Sakai, R, Swanson, G.T. | | Deposit date: | 2012-01-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of a Tetrameric Galectin from Cinachyrella Sp. (Ball Sponge).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ISU
 
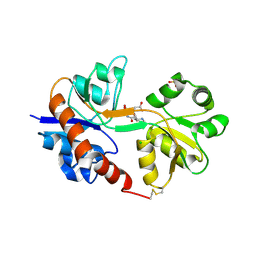 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
3GBA
 
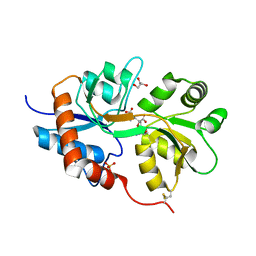 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
3GBB
 
 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with MSVIII-19 at 2.10A resolution | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, Glutamate receptor, ionotropic kainate 1 | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
