1JIE
 
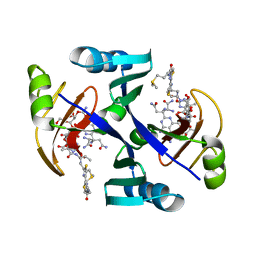 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with metal-free bleomycin | | Descriptor: | BLEOMYCIN A2, bleomycin-binding protein | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
1JIF
 
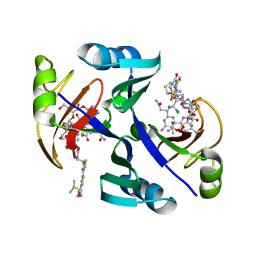 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with copper(II)-bleomycin | | Descriptor: | BLEOMYCIN A2, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
1IT5
 
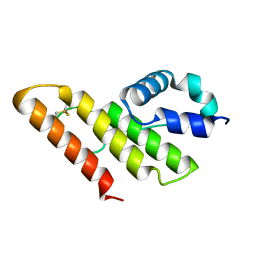 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
1NIQ
 
 | |
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1QTO
 
 | | 1.5 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT FROM BLEOMYCIN-PRODUCING STREPTOMYCES VERTICILLUS | | Descriptor: | BLEOMYCIN-BINDING PROTEIN | | Authors: | Kawano, Y, Kumagai, T, Muta, K, Matoba, Y, Davies, J, Sugiyama, M. | | Deposit date: | 1999-06-28 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a bleomycin resistance determinant from bleomycin-producing Streptomyces verticillus.
J.Mol.Biol., 295, 2000
|
|
1EWJ
 
 | | CRYSTAL STRUCTURE OF BLEOMYCIN-BINDING PROTEIN COMPLEXED WITH BLEOMYCIN | | Descriptor: | BLEOMYCIN A2, BLEOMYCIN RESISTANCE DETERMINANT | | Authors: | Maruyama, M, Kumagai, T, Matoba, Y, Hata, Y, Sugiyama, M. | | Deposit date: | 2000-04-26 | | Release date: | 2001-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1LWB
 
 | |
1ECS
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT ENCODED ON THE TRANSPOSON TN5 | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN, CALCIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Maruyama, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2000-01-25 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
1VFH
 
 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFT
 
 | | Crystal structure of L-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1KP4
 
 | |
6JIL
 
 | | Crystal structure of D-cycloserine synthetase DcsG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cycloserine biosynthesis protein DcsG, L(+)-TARTARIC ACID, ... | | Authors: | Matoba, Y, Sugiyama, M. | | Deposit date: | 2019-02-22 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Cyclization mechanism catalyzed by an ATP-grasp enzyme essential for d-cycloserine biosynthesis.
Febs J., 287, 2020
|
|
1MH6
 
 | | Solution Structure of the Transposon Tn5-encoding Bleomycin-binding Protein, BLMT | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN | | Authors: | Kumagai, T, Ohtani, K, Tsuboi, Y, Koike, T, Sugiyama, M. | | Deposit date: | 2002-08-19 | | Release date: | 2003-02-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transposon Tn5-encoding bleomycin-binding protein complexed with an activated bleomycin analogue.
To be published
|
|
1FAZ
 
 | |
5Y9N
 
 | | Crystal structure of Pyrococcus furiosus PbaA (monoclinic form), an archaeal homolog of proteasome-assembly chaperone | | Descriptor: | CHLORIDE ION, PbaA | | Authors: | Yagi-Utsumi, M, Sikdar, A, Kozai, T, Inoue, R, Sugiyama, M, Uchihashi, T, Satoh, T, Kato, K. | | Deposit date: | 2017-08-26 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conversion of functionally undefined homopentameric protein PbaA into a proteasome activator by mutational modification of its C-terminal segment conformation
Protein Eng. Des. Sel., 31, 2018
|
|
5B1H
 
 | |
5B1I
 
 | |
1WXC
 
 | | Crystal Structure of the copper-free Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein | | Descriptor: | MelC, NITRATE ION, tyrosinase | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
1WX5
 
 | | Crystal Structure of the copper-free Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein in the monoclinic crystal | | Descriptor: | CHLORIDE ION, MelC, SODIUM ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
1WX2
 
 | | Crystal Structure of the oxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein prepared by the addition of hydrogenperoxide | | Descriptor: | COPPER (II) ION, MelC, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
1WX4
 
 | | Crystal structure of the oxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase complexed with a caddie protein prepared by the addition of dithiothreitol | | Descriptor: | COPPER (II) ION, MelC, NITRATE ION, ... | | Authors: | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | Deposit date: | 2005-01-19 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
2A4W
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
2A4X
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Metal-Free Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
