3BZR
 
 | | Crystal structure of EscU C-terminal domain with N262D mutation, Space group P 41 21 2 | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZY
 
 | | Crystal structure of the mutated Y316D EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZP
 
 | | Crystal structural of the mutated N262A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
5WC3
 
 | | SpoIIIAG | | Descriptor: | SpoIIIAG, Stage III sporulation engulfment assemblyprotein | | Authors: | Zeytuni, N, Hong, C, Worrall, L.J, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1F00
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | Descriptor: | INTIMIN | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F02
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF INTIMIN IN COMPLEX WITH TRANSLOCATED INTIMIN RECEPTOR (TIR) INTIMIN-BINDING DOMAIN | | Descriptor: | INTIMIN, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-14 | | Release date: | 2000-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F6D
 
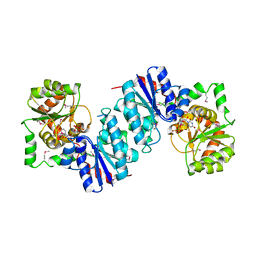 | | THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI. | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE, ... | | Authors: | Campbell, R.E, Mosimann, S.C, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases.
Biochemistry, 39, 2000
|
|
1FOF
 
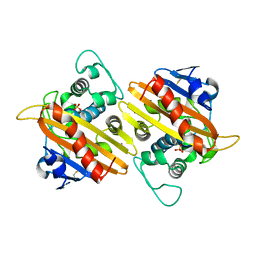 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 | | Descriptor: | BETA LACTAMASE OXA-10, COBALT (II) ION, SULFATE ION | | Authors: | Paetzel, M, Danel, F, de Castro, L, Mosimann, S.C, Page, M.G.P, Strynadka, N.C.J. | | Deposit date: | 2000-08-28 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the class D beta-lactamase OXA-10.
Nat.Struct.Biol., 7, 2000
|
|
1G68
 
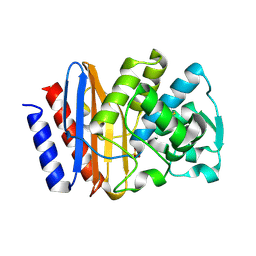 | | PSE-4 CARBENICILLINASE, WILD TYPE | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
1G6A
 
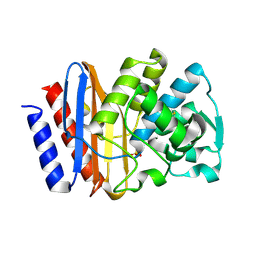 | | PSE-4 CARBENICILLINASE, R234K MUTANT | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
1G9R
 
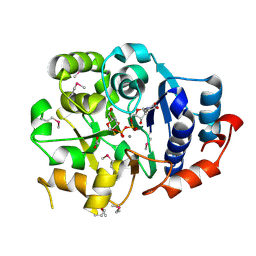 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GA8
 
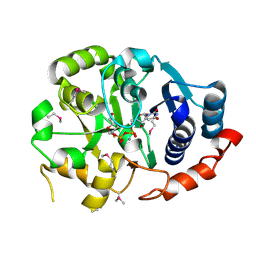 | | CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS. | | Descriptor: | 4-deoxy-beta-D-xylo-hexopyranose-(1-4)-beta-D-glucopyranose, GALACTOSYL TRANSFERASE LGTC, MANGANESE (II) ION, ... | | Authors: | Persson, K, Ly, H.D, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1JDI
 
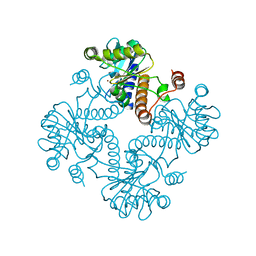 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | Descriptor: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | Authors: | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2001-06-13 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization.
Biochemistry, 40, 2001
|
|
1JHF
 
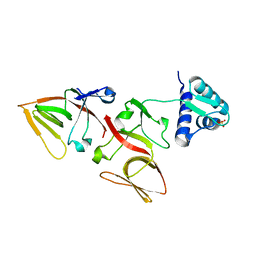 | | LEXA G85D MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHH
 
 | | LEXA S119A MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHC
 
 | | LEXA S119A C-TERMINAL TRYPTIC FRAGMENT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JTD
 
 | | Crystal structure of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase | | Descriptor: | CALCIUM ION, TEM-1 beta-lactamase, beta-lactamase inhibitor protein II | | Authors: | Lim, D.C, Park, H.U, De Castro, L, Kang, S.G, Lee, H.S, Jensen, S, Lee, K.J, Strynadka, N.C.J. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
1K3E
 
 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3S
 
 | | Type III Secretion Chaperone SigE | | Descriptor: | PHOSPHATE ION, SigE | | Authors: | Bertero, M.G, Luo, Y, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-28 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
