4S2O
 
 | | OXA-10 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, COBALT (II) ION | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2N
 
 | | OXA-48 in complex with Avibactam at pH 8.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2I
 
 | | CTX-M-15 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2K
 
 | | OXA-48 in complex with Avibactam at pH 7.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2P
 
 | |
2JW1
 
 | | Structural characterization of the type III pilotin-secretin interaction in Shigella flexneri by NMR spectroscopy | | Descriptor: | Lipoprotein mxiM, Outer membrane protein mxiD | | Authors: | Okon, M.S, Lario, P.I, Creagh, L, Jung, Y.M.T, Maurelli, A.T, Strynadka, N.C.J, McIntosh, L.P. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-02 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri
Structure, 16, 2008
|
|
2LQV
 
 | | YebF | | Descriptor: | Protein yebF | | Authors: | Prehna, G, Zhang, G, Gong, X, Duszyk, M, Okon, M, Mcintosh, L.P, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2012-03-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Protein Export Pathway Involving Escherichia coli Porins.
Structure, 20, 2012
|
|
2LV4
 
 | | ZirS C-terminal Domain | | Descriptor: | Putative outer membrane or exported protein | | Authors: | Prehna, G, Li, Y, Stoynov, N, Okon, M, Vukovic, M, Mcintosh, L.P, Foster, L.J, Finlay, B, Strynadka, N.C.J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The zinc regulated antivirulence pathway of salmonella is a multiprotein immunoglobulin adhesion system.
J.Biol.Chem., 287, 2012
|
|
2OBL
 
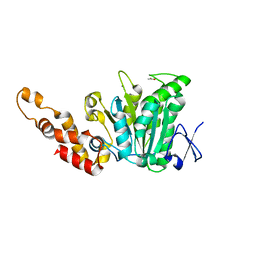 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ACETATE ION, CALCIUM ION, EscN, ... | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OBM
 
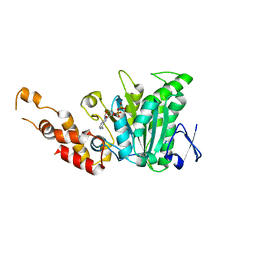 | | Structural and biochemical analysis of a prototypical ATPase from the type III secretion system of pathogenic bacteria | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, EscN | | Authors: | Zarivach, R, Vuckovic, M, Deng, W, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a prototypical ATPase from the type III secretion system.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2OLV
 
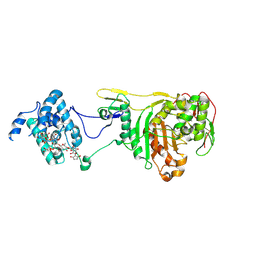 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L, Lim, D, Strynadka, N.C.J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2OYL
 
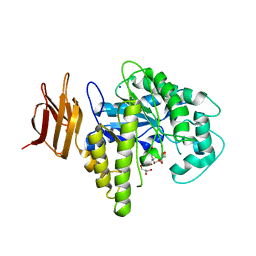 | | Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, Endoglycoceramidase II, GLYCEROL, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2OSY
 
 | |
2OYM
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex | | Descriptor: | Endoglycoceramidase II, N-{[(2R,3R,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PYRROLIDIN-2-YL]METHYL}-4-(DIMETHYLAMINO)BENZAMIDE, SODIUM ION | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2OSX
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | Descriptor: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
2OSW
 
 | | Endo-glycoceramidase II from Rhodococcus sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglycoceramidase II, SODIUM ION | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
2OYK
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, Endoglycoceramidase II, GLYCEROL, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3DWK
 
 | |
3ECQ
 
 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
3GMW
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Complex with TEM-1 Beta-Lactamase | | Descriptor: | B-lactamase, Beta-lactamase inhibitory protein BLIP-I, PHOSPHATE ION | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GR5
 
 | |
3GMX
 
 | |
3GR0
 
 | |
3GR1
 
 | |
3GMY
 
 | |
