1UXL
 
 | | I113T mutant of human SOD1 | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Hough, M.A, Grossmann, J.G, Antonyuk, S.V, Strange, R.W, Doucette, P.A, Rodriguez, J.A, Whitson, L.J, Hart, P.J, Hayward, L.J, Valentine, J.S, Hasnain, S.S. | | Deposit date: | 2004-02-25 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimer Destabilization in Superoxide Dismutase May Result in Disease-Causing Properties: Structures of Motor Neuron Disease Mutants
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3IWT
 
 | | Structure of hypothetical molybdenum cofactor biosynthesis protein B from Sulfolobus tokodaii | | Descriptor: | 178aa long hypothetical molybdenum cofactor biosynthesis protein B, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of hypothetical Mo-cofactor biosynthesis protein B (ST2315) from Sulfolobus tokodaii
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3KB6
 
 | | Crystal structure of D-Lactate dehydrogenase from aquifex aeolicus complexed with NAD and Lactic acid | | Descriptor: | D-lactate dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Ellis, M.J, Bessho, Y, Kuramitsu, S, Yokoyama, S, Hasnain, S.S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of D-lactate dehydrogenase from Aquifex aeolicus complexed with NAD(+) and lactic acid (or pyruvate).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6QWG
 
 | | Serial Femtosecond Crystallography Structure of Cu Nitrite Reductase from Achromobacter cycloclastes: Nitrite complex at Room Temperature | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A.E, Moreno-Chicano, T, Appleby, M.V, Worrall, J.W, Duyvesteyn, H.M.E, Strange, R.W, Beale, J, Axford, D, Sherrell, D.A, Sugimoto, H, Owada, S, Tono, K, Owen, R.L. | | Deposit date: | 2019-03-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6R2H
 
 | | Crystal structure of Apo PinO from Porphyromonas gingivitis | | Descriptor: | GLYCEROL, HmuY protein | | Authors: | Antonyuk, S.V, Bielecki, M, Strange, R.W, Capper, M, Olczak, T, Olczak, M. | | Deposit date: | 2019-03-17 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Prevotella intermedia produces two proteins homologous to Porphyromonas gingivalis HmuY but with different heme coordination mode.
Biochem.J., 477, 2020
|
|
5MJH
 
 | | X-ray generated oxyferrous/water mixed complex of DtpA from Streptomyces lividans | | Descriptor: | Dye type peroxidase A, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno Chicano, T, Chaplin, A.K, Worrall, J.A.R, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-12-01 | | Release date: | 2017-05-10 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction.
IUCrJ, 4, 2017
|
|
5MAP
 
 | | X-ray generated oxyferrous complex of DtpA from Streptomyces lividans | | Descriptor: | DtpA, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Moreno Chicano, T, Chaplin, A.K, Worrall, J.A.R, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photoreduction and validation of haem-ligand intermediate states in protein crystals by in situ single-crystal spectroscopy and diffraction.
IUCrJ, 4, 2017
|
|
3ZK4
 
 | |
4A7U
 
 | | Structure of human I113T SOD1 complexed with adrenaline in the p21 space group. | | Descriptor: | ACETATE ION, COPPER (II) ION, L-EPINEPHRINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7T
 
 | | Structure of human I113T SOD1 mutant complexed with isoproteranol in the p21 space group | | Descriptor: | ACETATE ION, COPPER (II) ION, ISOPRENALINE, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
4A7Q
 
 | | Structure of human I113T SOD1 mutant complexed with 4-(4-methyl-1,4- diazepan-1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-Ray Crystallography and Computational Docking for the Detection and Development of Protein-Ligand Interactions.
Curr.Med.Chem., 20, 2013
|
|
4A7G
 
 | | Structure of human I113T SOD1 mutant complexed with 4-methylpiperazin- 1-yl)quinazoline in the p21 space group. | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)QUINAZOLINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Sharma, R, Antonyuk, S.V, Strange, R.W, Berry, N.G, O'Neil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-24 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Curr.Med.Chem., 20, 2013
|
|
4A7S
 
 | | Structure of human I113T SOD1 mutant complexed with 5-Fluorouridine in the p21 space group | | Descriptor: | 5-FLUOROURIDINE, ACETATE ION, COPPER (II) ION, ... | | Authors: | Wright, G.S.A, Kershaw, N.M, Antonyuk, S.V, Strange, R.W, ONeil, P.M, Hasnain, S.S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-12-05 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Ligand Binding and Aggregation of Pathogenic Sod1.
Nat.Commun., 4, 2013
|
|
6ZU6
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUB
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 2 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUT
 
 | | Cu nitrite reductase MSOX series at 170K, dose point 5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Anyonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUD
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 3 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUA
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 4 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
7ADF
 
 | | SFX structure of dehaloperoxidase B in the ferric form | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
7ACP
 
 | | Serial synchrotron structure of dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
7ADX
 
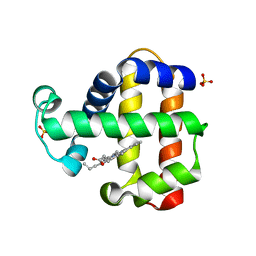 | | SFX structure of dehaloperoxidase B in the oxyferrous form | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moreno Chicano, T, Ebrahim, A, Worrall, J.W, Axford, D.A, Owada, S, Tosha, T, Sugimoto, H, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SFX structure of dehaloperoxidase B from Amphitrite ornata in the oxyferrous form
To Be Published
|
|
1OBF
 
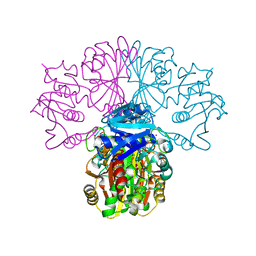 | | The crystal structure of Glyceraldehyde 3-phosphate Dehydrogenase from Alcaligenes xylosoxidans at 1.7A resolution. | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, POTASSIUM ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Eady, R.R, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Glyceraldehyde 3-Phosphate Dehydrogenase from Alcaligenes Xylosoxidans at 1.7 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HAU
 
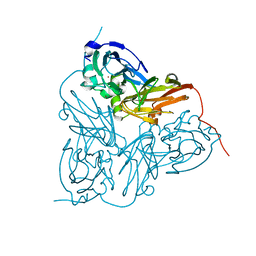 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HAW
 
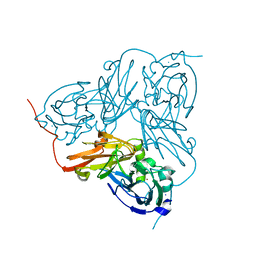 | | X-RAY STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE AT HIGH PH AND IN COPPER FREE FORM AT 1.9 A RESOLUTION | | Descriptor: | COPPER (I) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE | | Authors: | Ellis, M.J, Dodd, F.E, Strange, R.W, Prudencio, M, Sawerseady, R.R, Hasnain, S.S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of a Blue Copper Nitrite Reductase at High Ph and in Copper-Free Form at 1.9 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GS8
 
 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
