5WBF
 
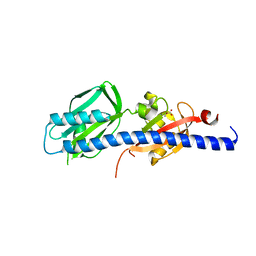 | | Double CACHE (dCACHE) sensing domain of TlpC chemoreceptor from Helicobacter pylori | | Descriptor: | GLYCEROL, LACTIC ACID, Methyl-accepting chemotaxis transducer (TlpC) | | Authors: | Machuca, M.A, Johnson, K.S, Liu, Y.C, Steer, D.L, Ottemann, K.M, Roujeinikova, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Helicobacter pylori chemoreceptor TlpC mediates chemotaxis to lactate.
Sci Rep, 7, 2017
|
|
3TI7
 
 | |
6PCA
 
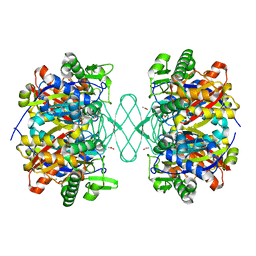 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
6PCB
 
 | |
6PCC
 
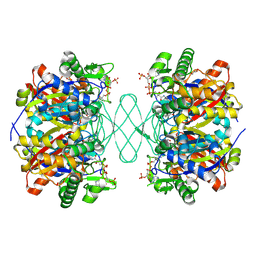 | |
6PCD
 
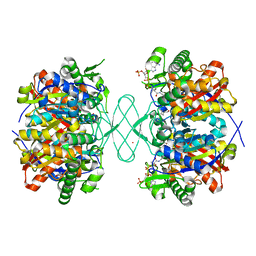 | |
8FX6
 
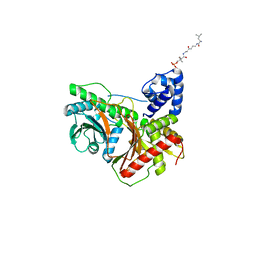 | | Non-ribosomal PCP-C didomain (amide stabilised leucine) acceptor bound state | | Descriptor: | N-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]-L-leucinamide, PCP-C didomain | | Authors: | Ho, Y.T.C, Cryle, M.J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Not always an innocent bystander: the impact of stabilised phosphopantetheine moieties when studying nonribosomal peptide biosynthesis.
Chem.Commun.(Camb.), 59, 2023
|
|
8FX7
 
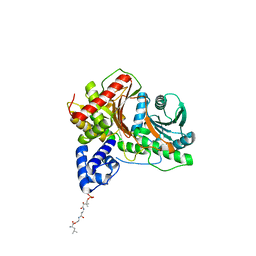 | | Non-ribosomal PCP-C didomain (ester stabilised leucine) acceptor bound state | | Descriptor: | 2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl L-leucinate, PCP-C didomain | | Authors: | Ho, Y.T.C, Cryle, M.J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Not always an innocent bystander: the impact of stabilised phosphopantetheine moieties when studying nonribosomal peptide biosynthesis.
Chem.Commun.(Camb.), 59, 2023
|
|
8G3J
 
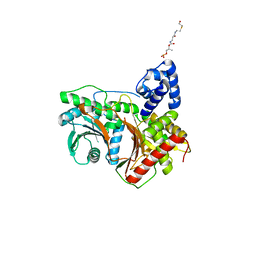 | |
8G3I
 
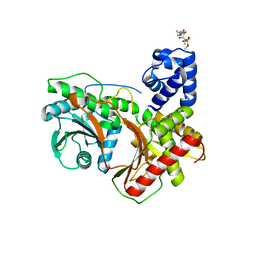 | |
3TI9
 
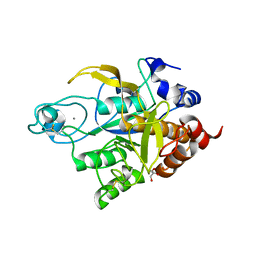 | | Crystal structure of the basic protease BprB from the ovine footrot pathogen, Dichelobacter nodosus | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong, W, Whisstock, J.C, Porter, C.J. | | Deposit date: | 2011-08-20 | | Release date: | 2011-10-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S1 Pocket of a Bacterially Derived Subtilisin-like Protease Underpins Effective Tissue Destruction.
J.Biol.Chem., 286, 2011
|
|
7KW0
 
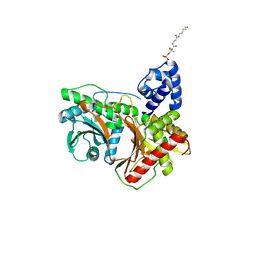 | | Non-ribosomal didomain (stabilised glycine-PCP-C) acceptor bound state | | Descriptor: | N-{2-[(2-aminoethyl)sulfanyl]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KVW
 
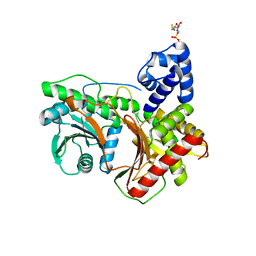 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KW2
 
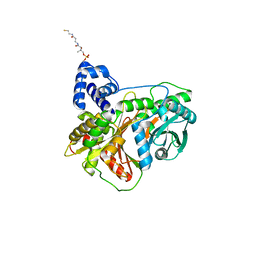 | | Non-ribosomal didomain (holo-PCP-C) acceptor bound state, R2577G | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PCP-C didomain | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
7KW3
 
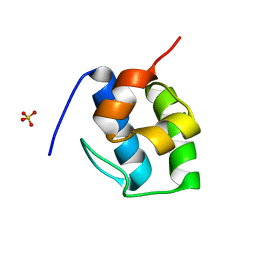 | | Non Ribosomal PCP domain | | Descriptor: | PCP domain, SULFATE ION | | Authors: | Izore, T, Ho, Y.T.C, Kaczmarski, J.A, Gavriilidou, A, Chow, K.H, Steer, D, Goode, R.J.A, Schittenhelm, R.B, Tailhades, J, Tosin, M, Challis, G.L, Krenske, E.H, Ziemert, N, Jackson, C.J, Cryle, M.J. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of a non-ribosomal peptide synthetase condensation domain suggest the basis of substrate selectivity.
Nat Commun, 12, 2021
|
|
