1BXL
 
 | | STRUCTURE OF BCL-XL/BAK PEPTIDE COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BAK PEPTIDE, BCL-XL | | Authors: | Sattler, M, Liang, H, Nettesheim, D, Meadows, R.P, Harlan, J.E, Eberstadt, M, Yoon, H, Shuker, S.B, Chang, B.S, Minn, A.J, Thompson, C.B, Fesik, S.W. | | Deposit date: | 1996-10-16 | | Release date: | 1997-10-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis.
Science, 275, 1997
|
|
2MJN
 
 | |
6R8G
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
1MHN
 
 | |
6DCL
 
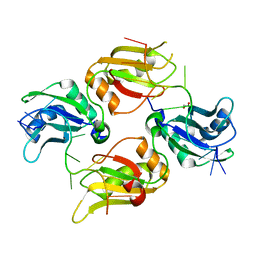 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
4UQT
 
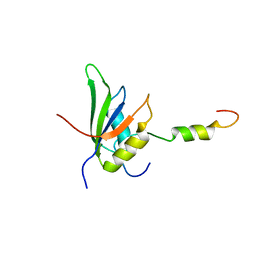 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
8POI
 
 | |
6R73
 
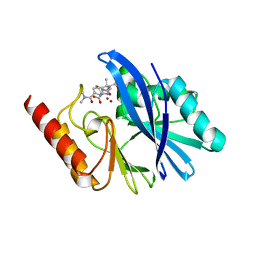 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R78
 
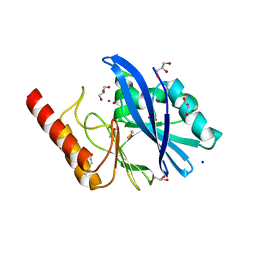 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R79
 
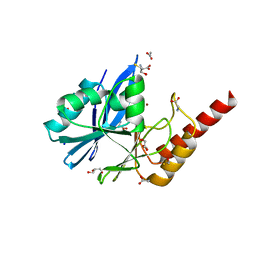 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop open) | | Descriptor: | BETA-MERCAPTOETHANOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
7Z0K
 
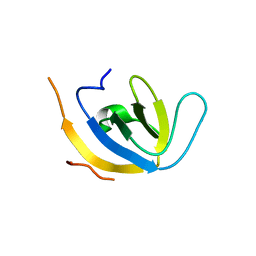 | |
7Z0J
 
 | | human PEX13 SH3 domain in complex with internal FxxxF motif | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13 | | Authors: | Gaussmann, S, Zak, K, Kreisz, N, Sattler, M. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
7Z0I
 
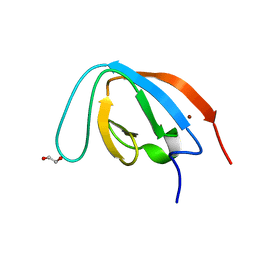 | | human PEX13 SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13, ZINC ION | | Authors: | Gaussmann, S, Zak, K, Sattler, M. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
8B8S
 
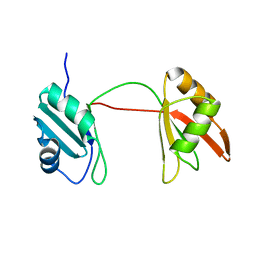 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-02-08 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
7NDX
 
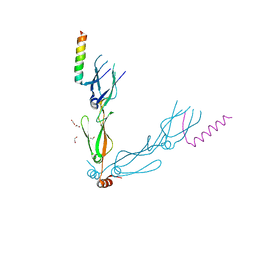 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
6ELD
 
 | |
8PXX
 
 | |
8PXW
 
 | |
1LXL
 
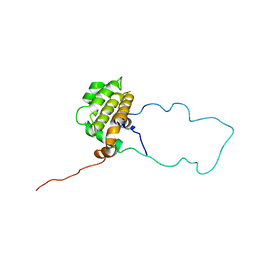 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1MAZ
 
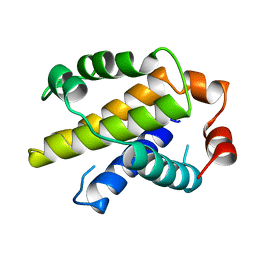 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
5OML
 
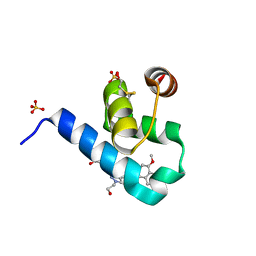 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope | | Descriptor: | (3~{R})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | Authors: | Ratkova, E.L, Dawidowski, M, Napolitano, V, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | Deposit date: | 2017-08-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope
To Be Published
|
|
1FHO
 
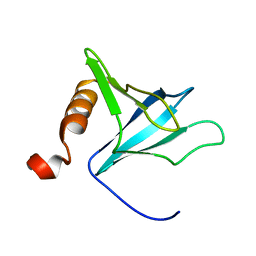 | | Solution Structure of the PH Domain from the C. Elegans Muscle Protein UNC-89 | | Descriptor: | UNC-89 | | Authors: | Blomberg, N, Baraldi, E, Sattler, M, Saraste, M, Nilges, M. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function.
Structure Fold.Des., 8, 2000
|
|
1D8B
 
 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
