1BJ7
 
 | | BOVINE LIPOCALIN ALLERGEN BOS D 2 | | Descriptor: | D 2 | | Authors: | Rouvinen, J. | | Deposit date: | 1998-07-02 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the molecular basis of allergy. three-dimensional structure of the bovine lipocalin allergen Bos d 2.
J.Biol.Chem., 274, 1999
|
|
3CBH
 
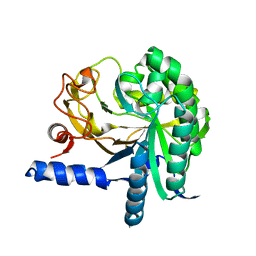 | |
1XYO
 
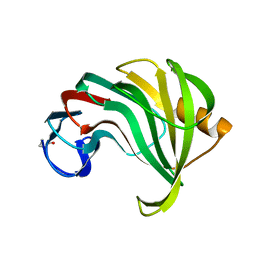 | |
1XYP
 
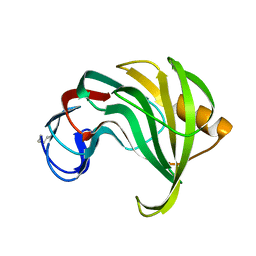 | |
1XYN
 
 | |
1RED
 
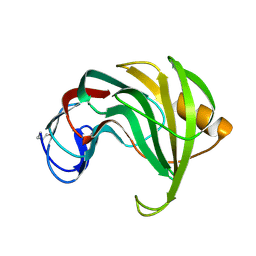 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 4,5-EPOXYPENTYL-BETA-D-XYLOSIDE | | Descriptor: | 3-[(2R)-oxiran-2-yl]propyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | Authors: | Rouvinen, J, Havukainen, R, Torronen, A. | | Deposit date: | 1995-12-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
1REF
 
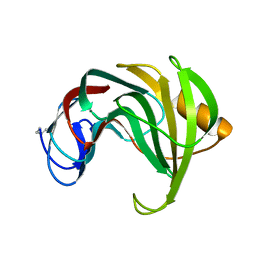 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 2,3-EPOXYPROPYL-BETA-D-XYLOSIDE | | Descriptor: | (2R)-oxiran-2-ylmethyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | Authors: | Rouvinen, J, Havukainen, R, Torronen, A. | | Deposit date: | 1995-12-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
1REE
 
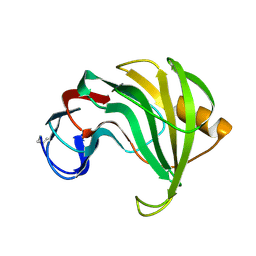 | | ENDO-1,4-BETA-XYLANASE II COMPLEX WITH 3,4-EPOXYBUTYL-BETA-D-XYLOSIDE | | Descriptor: | (3S)-3-hydroxybutyl beta-D-xylopyranoside, BENZOIC ACID, ENDO-1,4-BETA-XYLANASE II | | Authors: | Rouvinen, J, Havukainen, R, Torronen, A. | | Deposit date: | 1995-12-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Covalent binding of three epoxyalkyl xylosides to the active site of endo-1,4-xylanase II from Trichoderma reesei.
Biochemistry, 35, 1996
|
|
1ENX
 
 | |
1APY
 
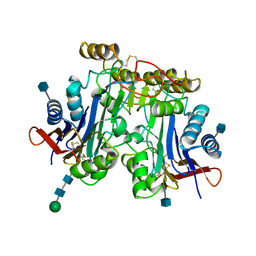 | | HUMAN ASPARTYLGLUCOSAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTYLGLUCOSAMINIDASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Rouvinen, J, Oinonen, C. | | Deposit date: | 1995-06-14 | | Release date: | 1996-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of human lysosomal aspartylglucosaminidase.
Nat.Struct.Biol., 2, 1995
|
|
1APZ
 
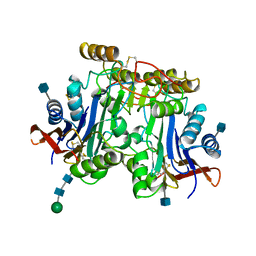 | |
3TTS
 
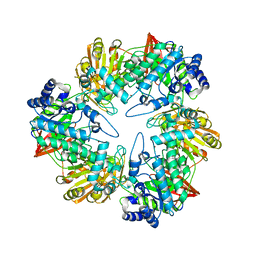 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
2RFZ
 
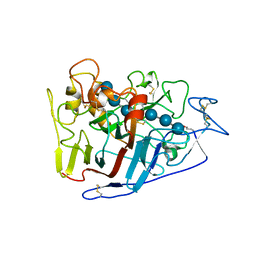 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellotriose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
1M4W
 
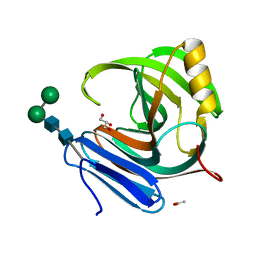 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
2FZ6
 
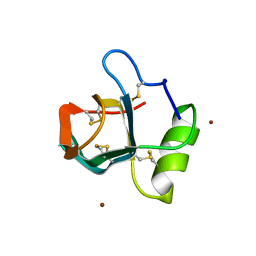 | | Crystal structure of hydrophobin HFBI | | Descriptor: | Hydrophobin-1, ZINC ION | | Authors: | Hakanpaa, J.M, Rouvinen, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Protein Sci., 15, 2006
|
|
5J83
 
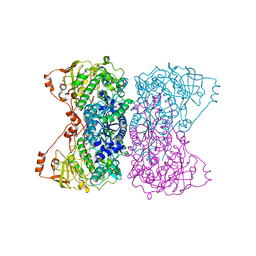 | |
5J85
 
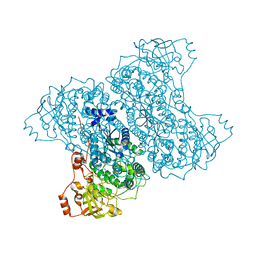 | | Ser480Ala mutant of L-arabinonate dehydratase | | Descriptor: | Dihydroxyacid dehydratase/phosphogluconate dehydratase, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2016-04-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of a Bacterial l-Arabinonate Dehydratase Contains a [2Fe-2S] Cluster.
ACS Chem. Biol., 12, 2017
|
|
5J84
 
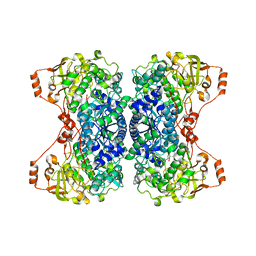 | |
3KDM
 
 | |
7PLC
 
 | | Caulobacter crescentus xylonolactonase with D-xylose, P21 space group | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
7PLD
 
 | |
7PLB
 
 | | Caulobacter crescentus xylonolactonase with D-xylose | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
6GSG
 
 | | Crystal structure of Aspergillus oryzae catechol oxidase complexed with resorcinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Penttinen, L, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Unraveling Substrate Specificity and Catalytic Promiscuity of Aspergillus oryzae Catechol Oxidase.
Chembiochem, 19, 2018
|
|
4UR7
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
