3EUJ
 
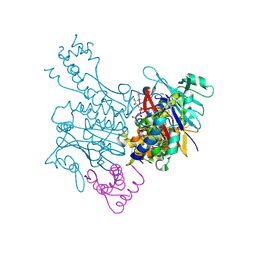 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, symmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukF, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUK
 
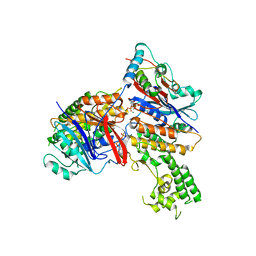 | | Crystal structure of MukE-MukF(residues 292-443)-MukB(head domain)-ATPgammaS complex, asymmetric dimer | | Descriptor: | Chromosome partition protein mukB, Linker, Chromosome partition protein mukE, ... | | Authors: | Woo, J.S, Lim, J.H, Shin, H.C, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3EUH
 
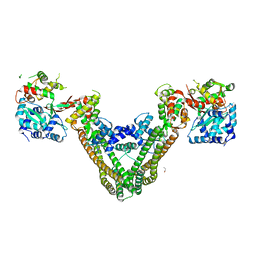 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
3FD9
 
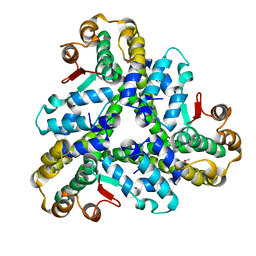 | |
3H77
 
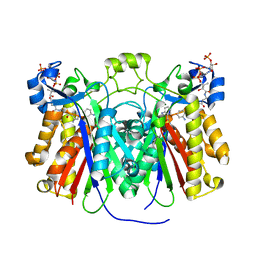 | | Crystal structure of Pseudomonas aeruginosa PqsD in a covalent complex with anthranilate | | Descriptor: | Anthraniloyl-coenzyme A, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3N0Y
 
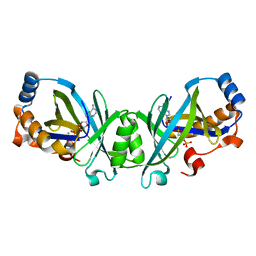 | | Adenylate cyclase class IV with active site ligand APC | | Descriptor: | Adenylate cyclase 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3N0Z
 
 | |
3H76
 
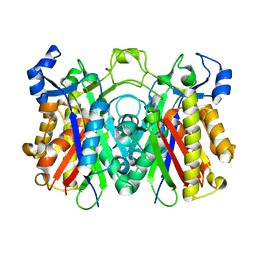 | | Crystal structure of PqsD, a key enzyme in Pseudomonas aeruginosa quinolone signal biosynthesis pathway | | Descriptor: | PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3H78
 
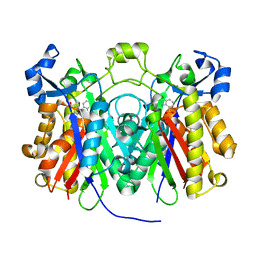 | | Crystal structure of Pseudomonas aeruginosa PqsD C112A mutant in complex with anthranilic acid | | Descriptor: | 2-AMINOBENZOIC ACID, PQS biosynthetic enzyme | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-04-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PqsD, a Pseudomonas quinolone signal biosynthetic enzyme, in complex with anthranilate.
Biochemistry, 48, 2009
|
|
3N10
 
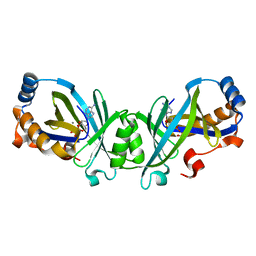 | | Product complex of adenylate cyclase class IV | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Adenylate cyclase 2, MANGANESE (II) ION, ... | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
3HGU
 
 | |
3HGV
 
 | |
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
2OIF
 
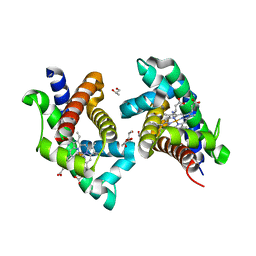 | |
4ZV0
 
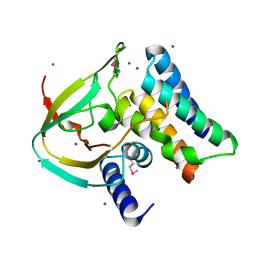 | | Structure of Tse6 in complex with Tsi6 | | Descriptor: | IODIDE ION, Tse6-binding/Tse6 immunity protein, antibacterial effector secreted protein (type VI secretion system) | | Authors: | Whitney, J.C, Sawai, S, Ralston, C, Mougous, J.D. | | Deposit date: | 2015-05-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | An Interbacterial NAD(P)(+) Glycohydrolase Toxin Requires Elongation Factor Tu for Delivery to Target Cells.
Cell, 163, 2015
|
|
4ZUA
 
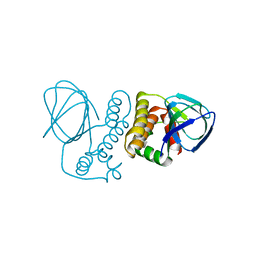 | | Crystal structure of the ExsA regulatory domain | | Descriptor: | Exoenzyme S synthesis regulatory protein ExsA | | Authors: | Schubot, F.D. | | Deposit date: | 2015-05-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of the Regulatory Domain of ExsA, a Key Transcriptional Regulator of the Type Three Secretion System in Pseudomonas aeruginosa.
Plos One, 10, 2015
|
|
3JRM
 
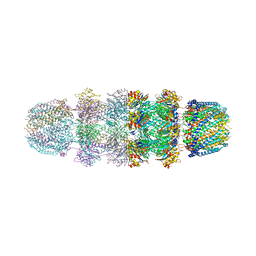 | |
3JSE
 
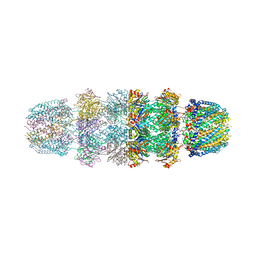 | |
2PWO
 
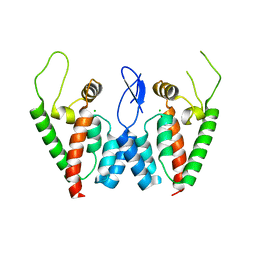 | | Crystal Structure of HIV-1 CA146 A92E Psuedo Cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol) | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2PWM
 
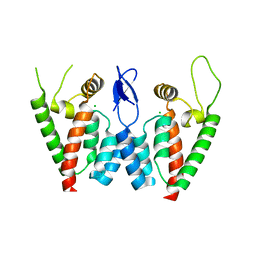 | | Crystal Structure of HIV-1 CA146 A92E real cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2PXR
 
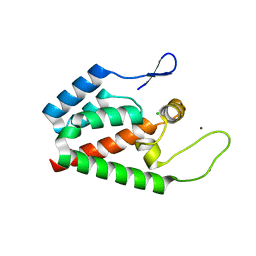 | | Crystal Structure of HIV-1 CA146 in the Presence of CAP-1 | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
3JTL
 
 | |
3PSJ
 
 | |
3PSI
 
 | |
3PSF
 
 | |
