3M3A
 
 | | The roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin: Cu(II)-I107E FeBMb (Cu(II) binding to FeB site) | | Descriptor: | COPPER (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M39
 
 | | The roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin: Fe(II)-I107E FeBMb (Fe(II) binding to FeB site) | | Descriptor: | FE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MC0
 
 | | Crystal Structure of Staphylococcal Enterotoxin G (SEG) in Complex with a Mouse T-cell Receptor beta Chain | | Descriptor: | ACETATE ION, Enterotoxin SEG, variable beta 8.2 mouse T cell receptor | | Authors: | Fernandez, M.M, Cho, S, Robinson, H, Mariuzza, R.A, Malchiodi, E.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of staphylococcal enterotoxin G (SEG) in complex with a mouse T-cell receptor {beta} chain.
J.Biol.Chem., 286, 2011
|
|
3M38
 
 | | The roles of Glutamates and Metal ions in a rationally designed nitric oxide reductase based on myoglobin: I107E FeBMb (No metal ion binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M3B
 
 | | The roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin: Zn(II)-I107E FeBMb (Zn(II) binding to FeB site) | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | Deposit date: | 2010-03-08 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MN0
 
 | | Introducing a 2-His-1-Glu Non-Heme Iron Center into Myoglobin confers Nitric Oxide Reductase activity: Cu(II)-CN-FeBMb(-His) form | | Descriptor: | COPPER (II) ION, CYANIDE ION, Myoglobin, ... | | Authors: | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Lei, L, Robinson, H, Lu, Y. | | Deposit date: | 2010-04-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Introducing a 2-his-1-glu nonheme iron center into myoglobin confers nitric oxide reductase activity.
J.Am.Chem.Soc., 132, 2010
|
|
2XS8
 
 | | Crystal Structure of ALIX in complex with the SIVagmTan-1 AYDPARKLL Late Domain | | Descriptor: | PROGRAMMED CELL DEATH 6-INTERACTING PROTEIN, SIVAGMTAN-1 GAG P6 | | Authors: | Zhai, Q, Landesman, M, Robinson, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Identification and Structural Characterization of the Alix-Binding Late Domains of Sivmac239 and Sivagmtan-1.
J.Virol., 85, 2011
|
|
2XS1
 
 | | Crystal Structure of ALIX in complex with the SIVmac239 PYKEVTEDL Late Domain | | Descriptor: | GAG POLYPROTEIN, PROGRAMMED CELL DEATH 6-INTERACTING PROTEIN | | Authors: | Zhai, Q, Landesman, M, Robinson, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Identification and Structural Characterization of the Alix-Binding Late Domains of Sivmac239 and Sivagmtan-1.
J.Virol., 85, 2011
|
|
2ZJG
 
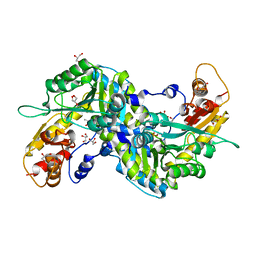 | | Crystal structural of mouse kynurenine aminotransferase III | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of mouse kynurenine aminotransferase III
To be Published
|
|
308D
 
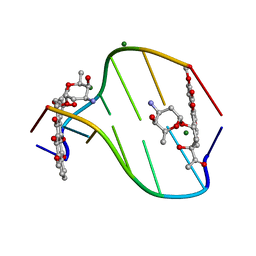 | | BINDING OF DAUNOMYCIN TO B-D-GLUCOSYLATED DNA FOUND IN PROTOZOA TRYPANOSOMA BRUCEI STUDIED BY X-RAY CRYSTALLOGRAPH | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Gao, Y.-G, Robinson, H, Wijsman, E.R, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1997-01-15 | | Release date: | 1997-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Daunomycin to beta-D-Glucosylated DNA Found in Protozoa Trypanosoma brucei Studied by X-Ray Crystallography
J.Am.Chem.Soc., 119, 1997
|
|
221D
 
 | |
222D
 
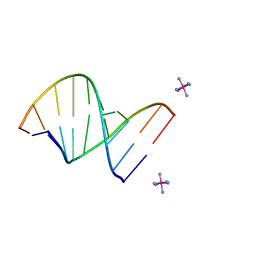 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA/RNA (5'-R(*GP*CP*)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
2B9C
 
 | | Structure of tropomyosin's mid-region: bending and binding sites for actin | | Descriptor: | striated-muscle alpha tropomyosin | | Authors: | Brown, J.H, Zhou, Z, Reshetnikova, L, Robinson, H, Yammani, R.D, Tobacman, L.S, Cohen, C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mid-region of tropomyosin: Bending and binding sites for actin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FJT
 
 | | Adenylyl cyclase class iv from Yersinia pestis | | Descriptor: | Adenylyl cyclase class IV, SULFATE ION | | Authors: | Gallagher, D.T, Smith, N.N, Kim, S.-K, Reddy, P.T, Robinson, H, Heroux, A. | | Deposit date: | 2006-01-03 | | Release date: | 2006-11-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the class IV adenylyl cyclase reveals a novel fold
J.Mol.Biol., 362, 2006
|
|
2F3L
 
 | |
2FZW
 
 | |
2G0Y
 
 | | Crystal Structure of a Lumenal Pentapeptide Repeat Protein from Cyanothece sp 51142 at 2.3 Angstrom Resolution. Tetragonal Crystal Form | | Descriptor: | CALCIUM ION, pentapeptide repeat protein | | Authors: | Kennedy, M.A, Ni, S, Buchko, G.W, Robinson, H. | | Deposit date: | 2006-02-13 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of two potentially universal turn motifs that shape the repeated five-residues fold - Crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142
Protein Sci., 15, 2006
|
|
2FZE
 
 | |
4WVP
 
 | | Crystal structure of an activity-based probe HNE complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BTN-3V3-NLB-OMT-OIC-3V2, ... | | Authors: | Lechtenberg, B.C, Kasperkiewicz, P, Robinson, H.R, Drag, M, Riedl, S.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Elastase-PK101 Structure: Mechanism of an Ultrasensitive Activity-based Probe Revealed.
Acs Chem.Biol., 10, 2015
|
|
4RT1
 
 | | Structure of the Alg44 PilZ domain (R95A mutant) from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44, CHLORIDE ION, ... | | Authors: | Whitfield, G.B, Whitney, J.C. | | Deposit date: | 2014-11-12 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
4RT0
 
 | | Structure of the Alg44 PilZ domain from Pseudomonas aeruginosa PAO1 in complex with c-di-GMP | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Alginate biosynthesis protein Alg44 | | Authors: | Whitfield, G.B, Whitney, J.C. | | Deposit date: | 2014-11-12 | | Release date: | 2015-04-08 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dimeric c-di-GMP Is Required for Post-translational Regulation of Alginate Production in Pseudomonas aeruginosa.
J.Biol.Chem., 290, 2015
|
|
5TSY
 
 | |
4FD6
 
 | |
4FD7
 
 | | Crystal structure of insect putative arylalkylamine N-Acetyltransferase 7 from the yellow fever mosquito Aedes aegypt | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, SULFATE ION, ... | | Authors: | Han, Q, Robinson, R, Li, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of insect arylalkylamine N-acetyltransferases: structural evidence from the yellow fever mosquito, Aedes aegypti.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FD5
 
 | |
