7VTG
 
 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) S30A mutant from Escherichia coli strain B | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89859128 Å) | | Cite: | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VVA
 
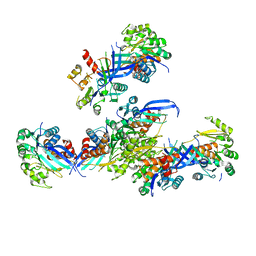 | | Pseudouridine bound structure of Pseudouridine kinase (PUKI) from Escherichia coli strain B | | Descriptor: | 5-[(2~{S},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1~{H}-pyrimidine-2,4-dione, Pseudouridine kinase | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75029182 Å) | | Cite: | Substrate-binding loop interactions with pseudouridine trigger conformational changes that promote catalytic efficiency of pseudouridine kinase PUKI.
J.Biol.Chem., 298, 2022
|
|
7VTD
 
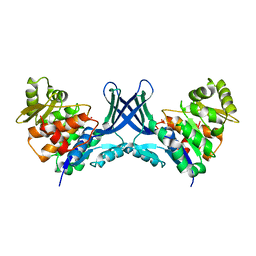 | |
7VTF
 
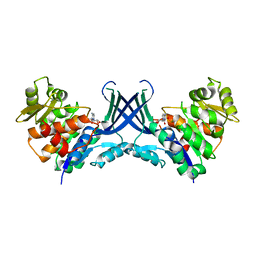 | |
7VTE
 
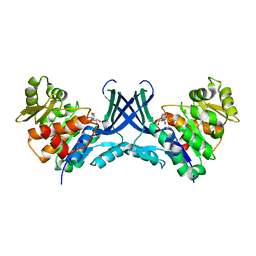 | |
7VRX
 
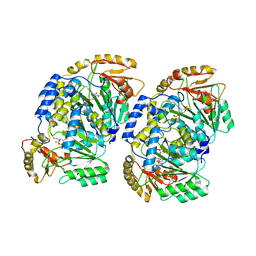 | | Pad-1 in the absence of substrate | | Descriptor: | Aminotransferase, SULFATE ION | | Authors: | Choi, M, Rhee, S. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96634674 Å) | | Cite: | Structural and biochemical basis for the substrate specificity of Pad-1, an indole-3-pyruvic acid aminotransferase in auxin homeostasis.
J.Struct.Biol., 214, 2022
|
|
4KL0
 
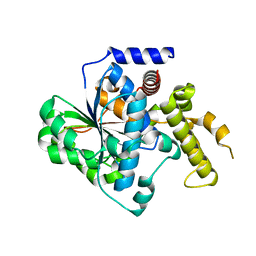 | | Crystal structure of the effector protein XOO4466 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of the effector protein XOO4466 from Xanthomonas oryzae
J.Struct.Biol., 184, 2013
|
|
5XU6
 
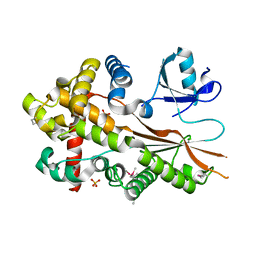 | | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase (IPK1) from Cryptococcus neoformans | | Descriptor: | Inositol-pentakisphosphate 2-kinase, SULFATE ION | | Authors: | Oh, J, Rhee, S. | | Deposit date: | 2017-06-22 | | Release date: | 2017-10-04 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from Cryptococcus neoformans.
J. Struct. Biol., 200, 2017
|
|
4PXB
 
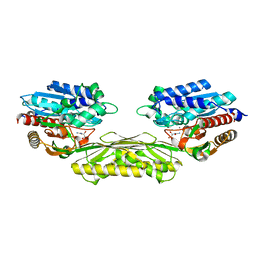 | | The crystal structure of AtUAH in complex with (S)-ureidoglycolate | | Descriptor: | (2S)-(carbamoylamino)(hydroxy)ethanoic acid, MANGANESE (II) ION, Ureidoglycolate hydrolase | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
4PXD
 
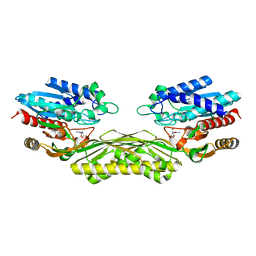 | | The crystal structure of EcAAH in complex with allantoate | | Descriptor: | ALLANTOATE ION, Allantoate amidohydrolase, MANGANESE (II) ION | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
4PXC
 
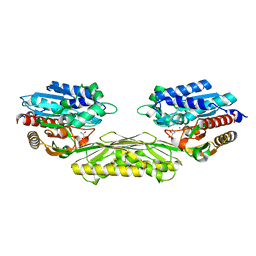 | | The crystal structure of AtUAH in complex with (S)-hydroxyglycine | | Descriptor: | (2S)-amino(hydroxy)ethanoic acid, MANGANESE (II) ION, Ureidoglycolate hydrolase | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
4PXE
 
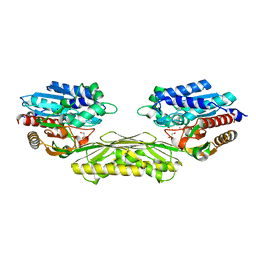 | | The crystal structure of AtUAH in complex with glyoxylate | | Descriptor: | GLYOXYLIC ACID, MANGANESE (II) ION, Ureidoglycolate hydrolase | | Authors: | Shin, I, Rhee, S. | | Deposit date: | 2014-03-23 | | Release date: | 2014-07-23 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J.Mol.Biol., 426, 2014
|
|
4RSX
 
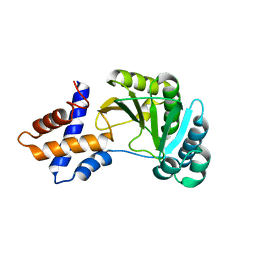 | |
4RSW
 
 | |
3DUL
 
 | |
3DUW
 
 | |
6ILT
 
 | | Structure of Arabidopsis thaliana Ribokinase complexed with ATP and Magnesium ion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribokinase, ... | | Authors: | Kang, P, Oh, J, Rhee, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analyses of ribokinase from Arabidopsis thaliana.
J. Struct. Biol., 206, 2019
|
|
6ILS
 
 | | Structure of Arabidopsis thaliana Ribokinase complexed with Ribose and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ribokinase, SODIUM ION, ... | | Authors: | Kang, P, Oh, J, Rhee, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mutational analyses of ribokinase from Arabidopsis thaliana.
J. Struct. Biol., 206, 2019
|
|
6ILR
 
 | |
3HQ0
 
 | | Crystal Structure Analysis of the 2,3-dioxygenase LapB from Pseudomonas in complex with a product | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohepta-2,4-dienoic acid, Catechol 2,3-dioxygenase, FE (III) ION | | Authors: | Cho, J.-H, Rhee, S. | | Deposit date: | 2009-06-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas
J.Biol.Chem., 284, 2009
|
|
3HPV
 
 | |
3HPY
 
 | |
6KIA
 
 | |
6KI9
 
 | | Apo structure of FabMG, novel types of Enoyl-acyl carrier protein reductase | | Descriptor: | 1,2-ETHANEDIOL, FabMG, novel types of Enoyl-acyl carrier protein reductase, ... | | Authors: | Kim, S, Rhee, S. | | Deposit date: | 2019-07-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclosan-resistance protein from the soil metagenome is a novel enoyl-acyl carrier protein reductase: Structure-guided functional analysis.
Febs J., 287, 2020
|
|
4FJS
 
 | | Crystal structure of ureidoglycolate dehydrogenase enzyme in apo form | | Descriptor: | Ureidoglycolate dehydrogenase | | Authors: | Kim, M.I, Shin, I, Lee, J, Rhee, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
