5YJJ
 
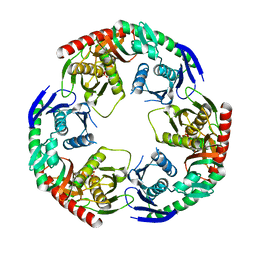 | | Crystal structure of PNPase from Staphylococcus epidermidis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Raj, R, Gopal, B. | | Deposit date: | 2017-10-10 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Staphylococcus epidermidis Polynucleotide phosphorylase and its interactions with ribonucleases RNase J1 and RNase J2.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6K6W
 
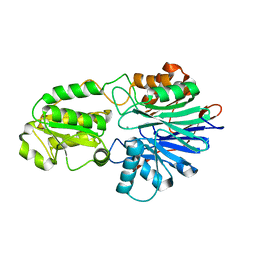 | |
6K6S
 
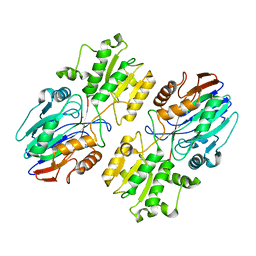 | |
8D5B
 
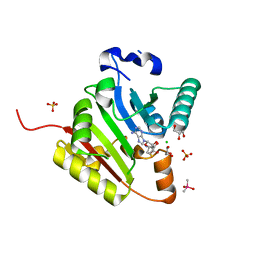 | | Crystal structure of human METTL1 in complex with SAH | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
8D58
 
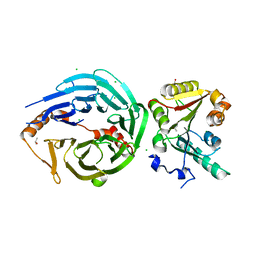 | | Crystal structure of human METTL1-WDR4 complex | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
8D59
 
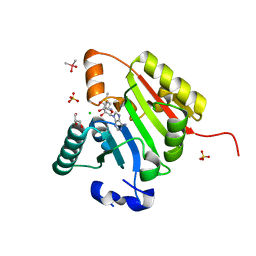 | | Crystal structure of human METTL1 in complex with SAM | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raj, R, Babu, K, Nam, Y. | | Deposit date: | 2022-06-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanisms of tRNA methylation by METTL1-WDR4.
Nature, 613, 2023
|
|
6WQ1
 
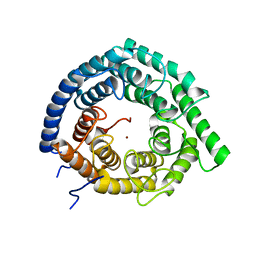 | | Eukaryotic LanCL2 protein | | Descriptor: | LanC-like protein 2, ZINC ION | | Authors: | Nair, S.K, Garg, N. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome.
Cell, 184, 2021
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
6T35
 
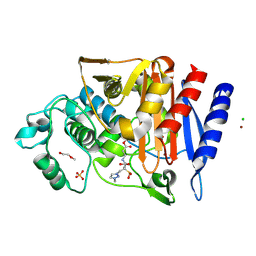 | | Crystal structure of AmpC from E.coli with Enmetazobactam (AAI-101) | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3U
 
 | | OXA-10 beta-lactamase with covalent modification | | Descriptor: | Beta-lactamase OXA-10, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3S
 
 | | OXA-10 beta-lactamase with S67Dha modification | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SODIUM ION, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-12-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Studies on enmetazobactam clarify mechanisms of widely used beta-lactamase inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7B3R
 
 | |
8D9K
 
 | |
8D9L
 
 | |
8EG0
 
 | |
