6QKU
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Chloroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKW
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKT
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 24hr - Glycolate bound | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKS
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
5JQH
 
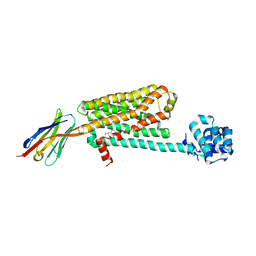 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
3KJ6
 
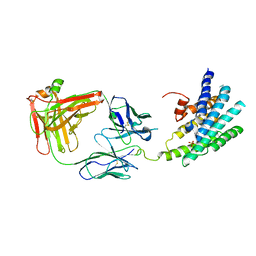 | | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex | | Descriptor: | Beta-2 adrenergic receptor, Fab heavy chain, Fab light chain, ... | | Authors: | Bokoch, M.P, Zou, Y, Rasmussen, S.G.F, Liu, C.W, Nygaard, R, Rosenbaum, D.M, Fung, J.J, Choi, H.-J, Thian, F.S, Kobilka, T.S, Puglisi, J.D, Weis, W.I, Pardo, L, Prosser, S, Mueller, L, Kobilka, B.K. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor.
Nature, 463, 2010
|
|
5T4T
 
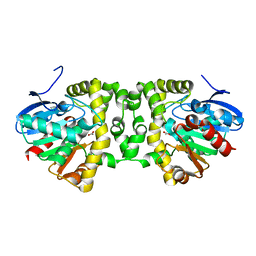 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn - Apo No Halide | | Descriptor: | ACETATE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5SWN
 
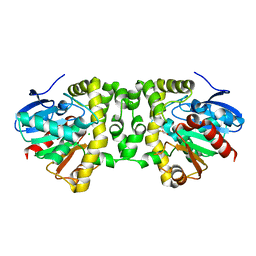 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3B
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3A
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3D
 
 | |
5K3C
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/5-Fluorotryptophan | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3F
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
