7QX0
 
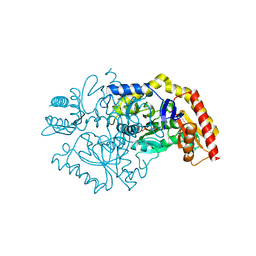 | | Transaminase Structure of Plurienzyme (Tr2E2) in complex with PLP | | Descriptor: | Aminotransferase TR2, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QX3
 
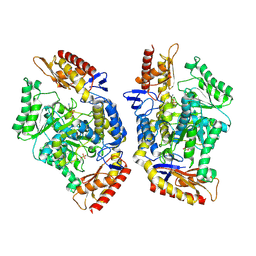 | | Structure of the transaminase TR2E2 with EOS | | Descriptor: | 2-azanylethyl hydrogen sulfate, Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7QYF
 
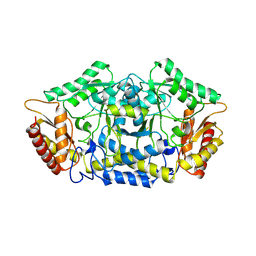 | | Structure of the transaminase PluriZyme variant (TR2E2) | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QYG
 
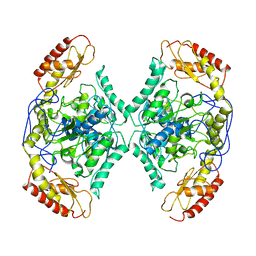 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
3U75
 
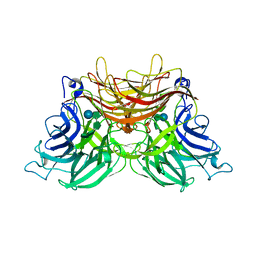 | | Structure of E230A-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosylnystose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
6EPB
 
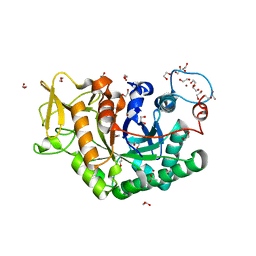 | | Structure of Chitinase 42 from Trichoderma harzianum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endochitinase 42, ... | | Authors: | Ramirez-Escudero, M, Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity.
Microb. Cell Fact., 17, 2018
|
|
6RB0
 
 | |
8PC7
 
 | |
6I8F
 
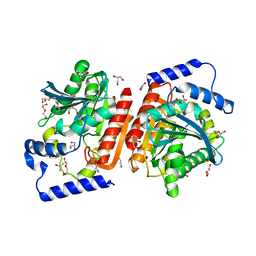 | |
3U14
 
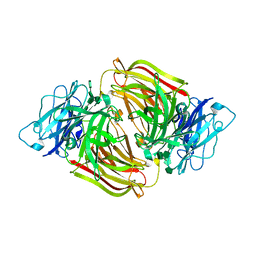 | | Structure of D50A-fructofuranosidase from Schwanniomyces occidentalis complexed with inulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fructofuranosidase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and kinetic insights reveal that the amino acid pair GLN228/ASN254 modulates the transfructosylating specificity of Schwanniomyces occidentalis beta-fructofuranosidase, an enzyme that produces prebiotics.
J.Biol.Chem., 287, 2012
|
|
5O47
 
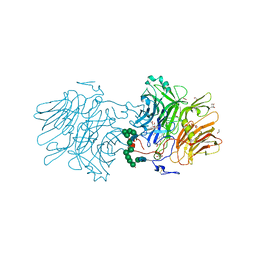 | |
5NSL
 
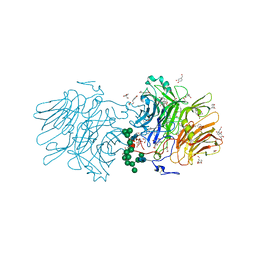 | |
6FJG
 
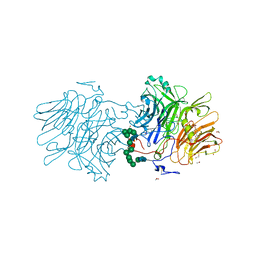 | |
6FJE
 
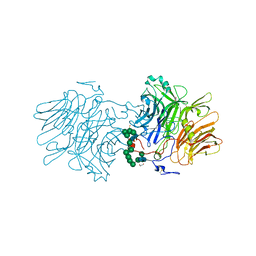 | |
7PP8
 
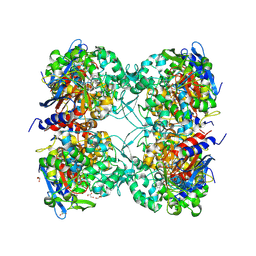 | |
7PP3
 
 | |
7PU6
 
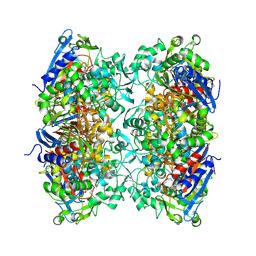 | |
8BET
 
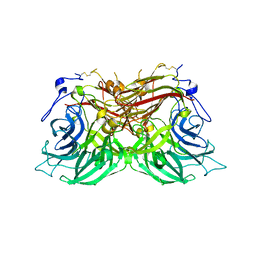 | |
8BEU
 
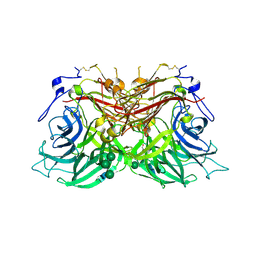 | |
8BES
 
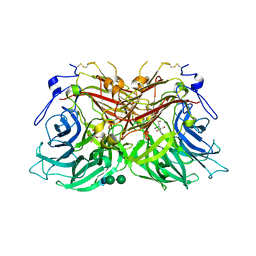 | |
8BEQ
 
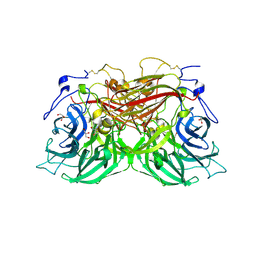 | | Structure of fructofuranosidase from Rhodotorula dairenensis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential.
Int J Mol Sci, 23, 2022
|
|
6RKY
 
 | |
6S82
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Tris-buffer molecule And hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
6S2H
 
 | |
6S2G
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Fructose And Epigallocatechin Gallate (Egcg) | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
