7SYJ
 
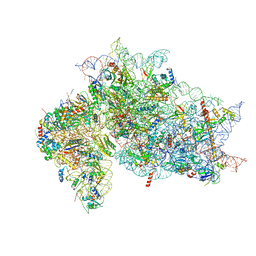 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYW
 
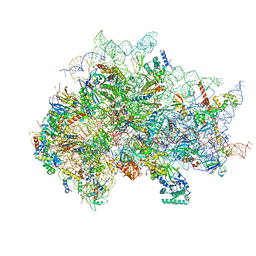 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYT
 
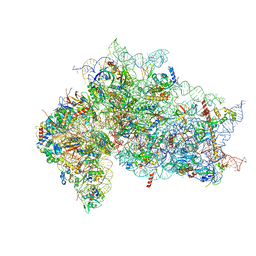 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6ZVK
 
 | | The Halastavi arva virus (HalV) intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES17, ... | | Authors: | Abaeva, I.S, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
3E1Y
 
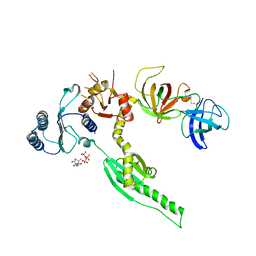 | | Crystal structure of human eRF1/eRF3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
3E20
 
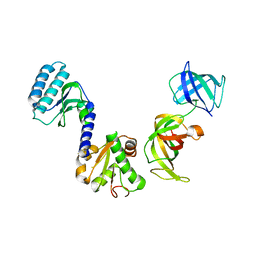 | | Crystal structure of S.pombe eRF1/eRF3 complex | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, Eukaryotic peptide chain release factor subunit 1 | | Authors: | Cheng, Z, Lim, M, Kong, C, Song, H. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into eRF3 and stop codon recognition by eRF1
Genes Dev., 23, 2009
|
|
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
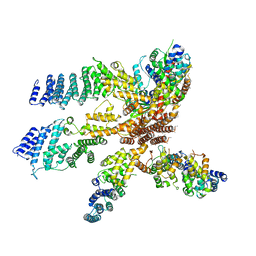 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
