4XDA
 
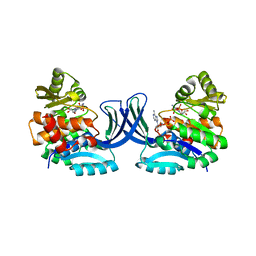 | | Vibrio cholerae O395 Ribokinase complexed with Ribose, ADP and Sodium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribokinase, SODIUM ION, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation
Adv.Exp.Med.Biol., 842, 2015
|
|
4XCK
 
 | | Vibrio cholerae O395 Ribokinase complexed with ADP, Ribose and Cesium ion. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CESIUM ION, Ribokinase, ... | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of Apo and Ligand Bound Vibrio cholerae Ribokinase (Vc-RK): Role of Monovalent Cation Induced Activation and Structural Flexibility in Sugar Phosphorylation.
Adv. Exp. Med. Biol., 842, 2015
|
|
4X8F
 
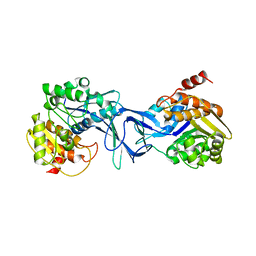 | | Vibrio cholerae O395 Ribokinase in apo form | | Descriptor: | Ribokinase | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Apo and Ligand
Bound Vibrio choleraeRibokinase (Vc-RK):
Role of Monovalent Cation Induced Activation
and Structural Flexibility in Sugar
Phosphorylation
Adv Exp Med Biol., 842, 2015
|
|
5EY7
 
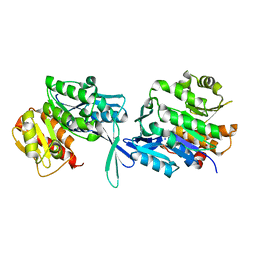 | |
5F11
 
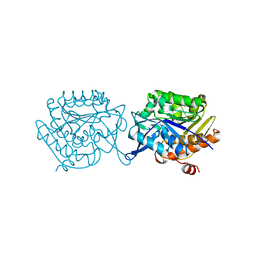 | |
5EYN
 
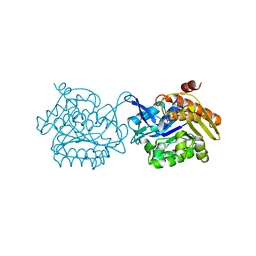 | | Crystal structure of Fructokinase from Vibrio cholerae O395 in fructose, ADP, Beryllium trifluoride and calcium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Paul, R, Nath, S, Sen, U. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.289 Å) | | Cite: | Crystal structure of Fructokinase from Vibrio cholerae O395 in apo form
To Be Published
|
|
5F0Z
 
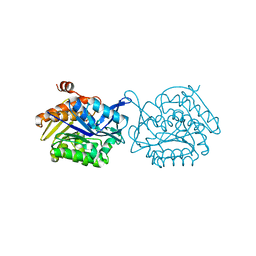 | |
5YGG
 
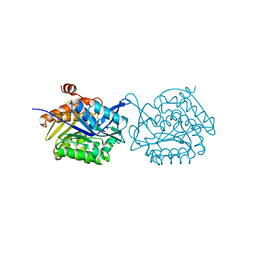 | | Crystal structure of Fructokinase Double-Mutant (T261C-H108C) from Vibrio cholerae O395 in fructose, ADP and potassium ion bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Fructokinase, ... | | Authors: | Chatterjee, S, Paul, R, Nath, S, Sen, U. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | Large-scale conformational changes and redistribution of surface negative charge upon sugar binding dictate the fidelity of phosphorylation in Vibrio cholerae fructokinase.
Sci Rep, 8, 2018
|
|
1FUE
 
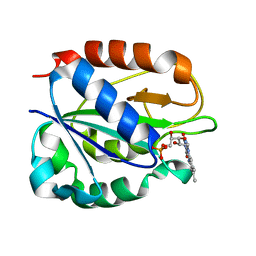 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
1W25
 
 | | Response regulator PleD in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, STALKED-CELL DIFFERENTIATION CONTROLLING PROTEIN, ... | | Authors: | Chan, C, Schirmer, T, Jenal, U. | | Deposit date: | 2004-06-28 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Activity and Allosteric Control of Diguanylate Cyclase
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2V0N
 
 | | ACTIVATED RESPONSE REGULATOR PLED IN COMPLEX WITH C-DIGMP AND GTP- ALPHA-S | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | Authors: | Wassmann, P, Schirmer, T. | | Deposit date: | 2007-05-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of Bef3--Modified Response Regulator Pled: Implications for Diguanylate Cyclase Activation, Catalysis, and Feedback Inhibition
Structure, 15, 2007
|
|
8P1V
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 6-cyclopropyl-~{N}-(2-methylindazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
8P4H
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric compound IDEAYA cmpd A | | Descriptor: | 7-chloranyl-4-[(3R)-3-fluoranylpyrrolidin-1-yl]-1-phenyl-quinazolin-2-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
8P1W
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with allosteric compound STL232591 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 8-methoxy-1-(4-methoxyphenyl)-3-methyl-2-oxidanyl-[1]benzofuro[3,2-c]pyridine, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
8P1T
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Z237451470 | | Descriptor: | 1,2-ETHANEDIOL, 6-cyclopropyl-~{N}-(1~{H}-indazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
