2G8J
 
 | | Calpain 1 proteolytic core in complex with SNJ-1945, a alpha-ketoamide-type inhibitor. | | Descriptor: | ((1S)-1-((((1S)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2,3-DIOXOPROPYL)AMINO)CARBONYL)-3-METHYLBUTYL)CARBAMIC ACID 5-METHOXY-3-OXAPENTYL ESTER, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
2G8E
 
 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
4MF3
 
 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
2GBL
 
 | | Crystal Structure of Full Length Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.R, Pattanayek, S, Xu, Y, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2006-03-10 | | Release date: | 2007-01-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods.
Embo J., 25, 2006
|
|
2HDA
 
 | | Yes SH3 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Yes, SULFATE ION | | Authors: | Camara-Artigas, A, Luque, I, Ruiz-Sanz, J, Mateo, P.L, Martin-Garcia, J.M. | | Deposit date: | 2006-06-20 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of the SH3 domain of the human c-Yes tyrosine kinase: Loop flexibility and amyloid aggregation.
Febs Lett., 581, 2007
|
|
2GRK
 
 | |
5MOB
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, SULFATE ION, SlPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MMX
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CSPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Moreno-Alvero, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MND
 
 | |
5MOA
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | SlPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MMQ
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | CSPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MN0
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CHLORIDE ION, CSPYL1, ... | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MSI
 
 | |
1TEW
 
 | |
1EWW
 
 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
6LA8
 
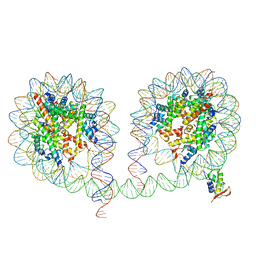 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6LA9
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6L9Z
 
 | | 338 bp di-nucleosome assembled with linker histone H1.X | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (338-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LAB
 
 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LER
 
 | | 169 bp nucleosome harboring non-identical cohesive DNA termini. | | Descriptor: | CALCIUM ION, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Sharma, D, Adhireksan, Z, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-26 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6M3V
 
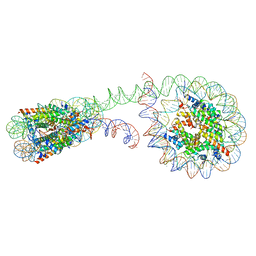 | | 355 bp di-nucleosome harboring cohesive DNA termini | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6M44
 
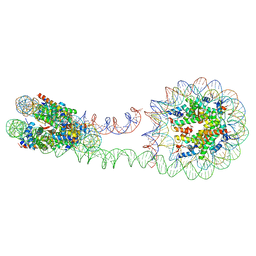 | | 355 bp di-nucleosome harboring cohesive DNA termini (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
