6KE9
 
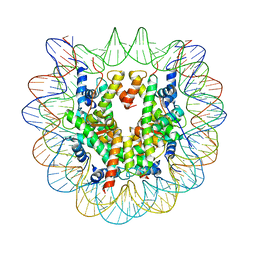 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-07-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6LE9
 
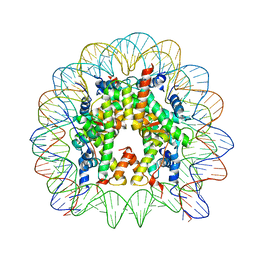 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-24 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6L9H
 
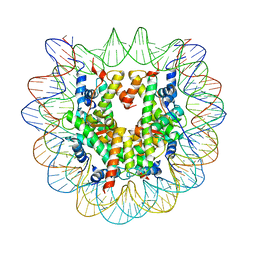 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6M9B
 
 | |
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAB
 
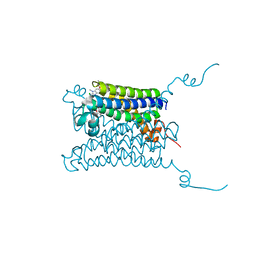 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4X29
 
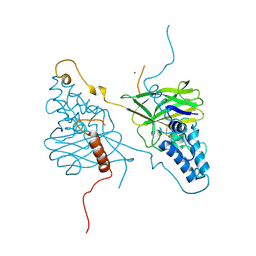 | |
4X27
 
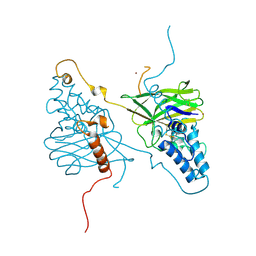 | |
7EFT
 
 | |
4YN2
 
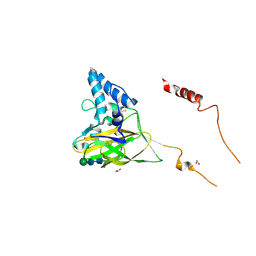 | | THE ATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, FUSOLIN, ZINC ION, ... | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TPZ
 
 | |
3PYI
 
 | |
3ZEK
 
 | |
3ZEJ
 
 | |
6J4P
 
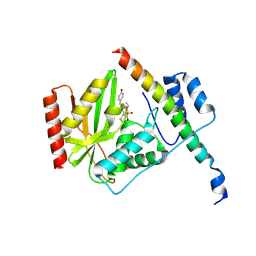 | |
6J4V
 
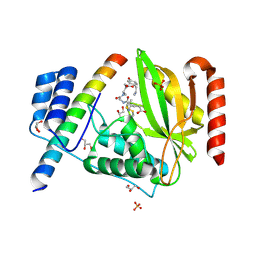 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4U
 
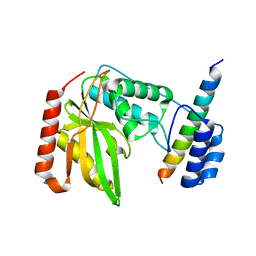 | |
6J4Q
 
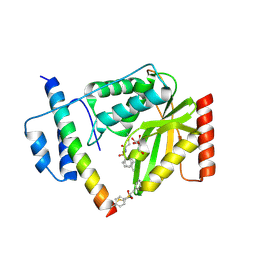 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J4O
 
 | |
6J4S
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4TO7
 
 | |
4TQ7
 
 | |
4TTZ
 
 | |
4TTW
 
 | |
4TTX
 
 | |
