7ONG
 
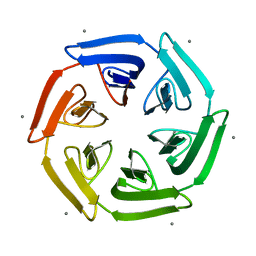 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | Descriptor: | CALCIUM ION, SAKe6BE-L1 | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONE
 
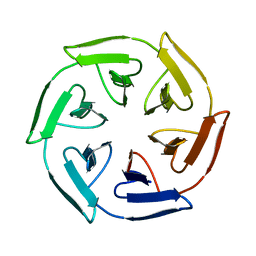 | | Crystal structure of the self-assembled SAKe6BE designer protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ON7
 
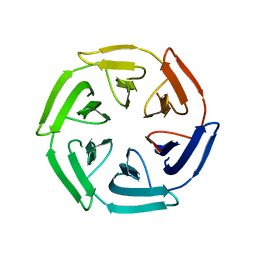 | |
7ON6
 
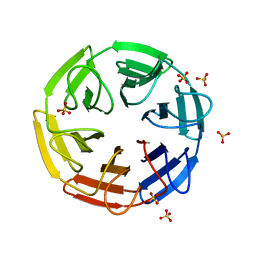 | | Crystal structure of the computationally designed SAKe6AE protein | | Descriptor: | SAKe6AE, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONH
 
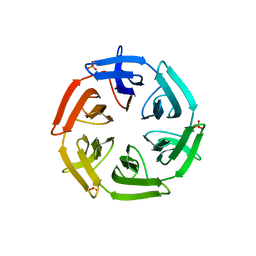 | | Crystal structure of the computationally designed SAKe6BE-L3 protein | | Descriptor: | SAKe6BE-L3, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OP4
 
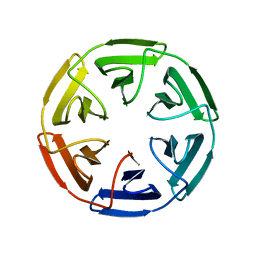 | |
7OPU
 
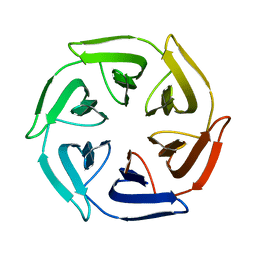 | |
7OV7
 
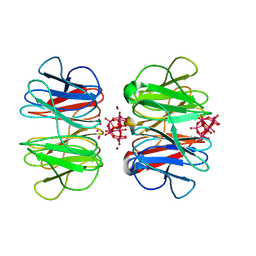 | | The hybrid cage formed between Pizza6-S and Cu(II)-substituted trilacunary Keggin | | Descriptor: | COPPER (II) ION, POTASSIUM ION, Pizza6-S, ... | | Authors: | Vandebroek, L, Noguchi, H, Anyushin, A, Van Meervelt, L, Voet, A.R.D, Parac-Vogt, T.N. | | Deposit date: | 2021-06-14 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hierarchical Self-Assembly of a Supramolecular Protein-Metal Cage Encapsulating a Polyoxometalate Guest
Cryst.Growth Des., 2022
|
|
7OPV
 
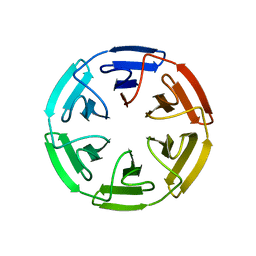 | | Crystal structure of the computationally designed SAKe6BE-3HH protein, alternative packing | | Descriptor: | SAKe6BE-3HH | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7PVP
 
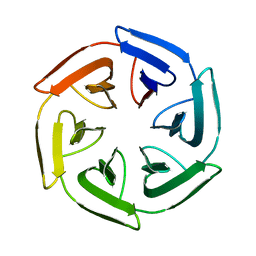 | | Crystal structure of the SAKe6BC designer protein | | Descriptor: | SAKe6BC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-10-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7Q58
 
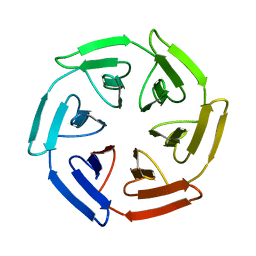 | | Crystal structure of the SAKe6BR designer protein | | Descriptor: | SAKe6BR | | Authors: | Wouters, S.M.L, Noguchi, H, Velupla, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-11-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
2D3M
 
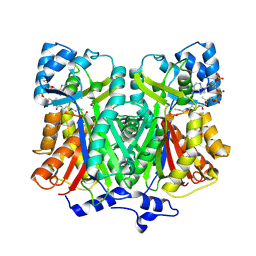 | | Pentaketide chromone synthase complexed with coenzyme A | | Descriptor: | COENZYME A, pentaketide chromone synthase | | Authors: | Morita, H, Kondo, S, Oguro, S, Noguchi, H, Sugio, S, Abe, I, Kohno, T. | | Deposit date: | 2005-09-29 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insight into Chain-Length Control and Product Specificity of Pentaketide Chromone Synthase from Aloe arborescens
Chem.Biol., 14, 2007
|
|
6I3B
 
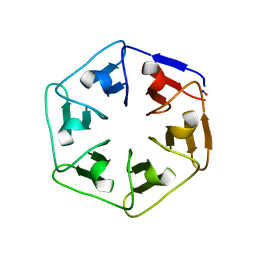 | |
6I37
 
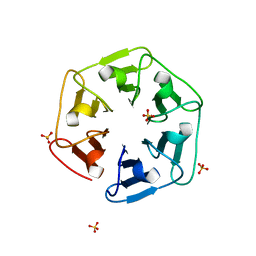 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | SULFATE ION, nv1Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | MAGNESIUM ION, v31Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I38
 
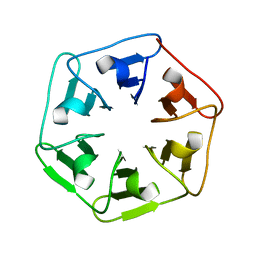 | |
6I3A
 
 | | Crystal structure of v22Pizza6-AYW, a circularly permuted designer protein | | Descriptor: | BROMIDE ION, v22Pizza6-AYW | | Authors: | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
4ZCN
 
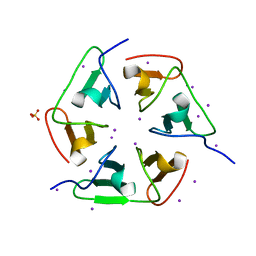 | | Crystal structure of nvPizza2-S16S58 | | Descriptor: | IODIDE ION, SULFATE ION, nvPizza2-S16S58 | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of nvPizza2-S16S58
To Be Published
|
|
5CHB
 
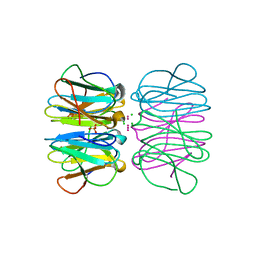 | | Crystal structure of nvPizza2-S16H58 coordinating a CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2015-07-10 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3VUS
 
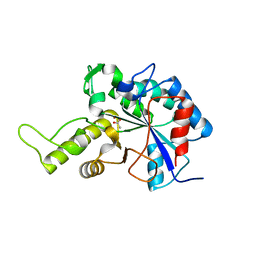 | | Escherichia coli PgaB N-terminal domain | | Descriptor: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-07-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WWF
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | pizza2sr-pb protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WWB
 
 | | Crystal structure of the computationally designed Pizza2-SR protein | | Descriptor: | Pizza2-SR protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WMV
 
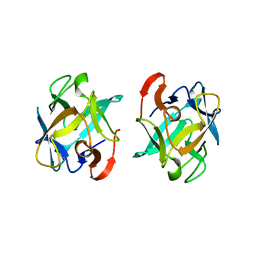 | | The structure of an anti-cancer lectin mytilec with ligand from the mussel Mytilus galloprovincialis | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
3WWA
 
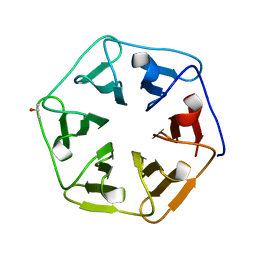 | | Crystal structure of the computationally designed Pizza7 protein after heat treatment | | Descriptor: | ISOPROPYL ALCOHOL, Pizza7H protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WW9
 
 | | Crystal structure of the computationally designed Pizza6 protein | | Descriptor: | GLYCEROL, SULFATE ION, pizza6 protein | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
