7P4A
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression. | | Descriptor: | Sri, Stl | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Tan, J.A, Cogdell, R.J, Chen, J, Byron, O, Christie, G.E, Marina, A, Penades, J.R. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
7ZVI
 
 | | Non-canonical Staphylococcus aureus pathogenicity island repression | | Descriptor: | Orf22, Sri | | Authors: | Miguel-Romero, L, Alqasmi, M, Bacarizo, J, Marina, A, Penades, J.R. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.973 Å) | | Cite: | Non-canonical Staphylococcus aureus pathogenicity island repression.
Nucleic Acids Res., 50, 2022
|
|
5LWK
 
 | | MaeR response regulator bound to beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein | | Authors: | Miguel-Romero, L, Casino, P, Landete, J.M, Monedero, V, Zuniga, M, Marina, A. | | Deposit date: | 2016-09-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The malate sensing two-component system MaeKR is a non-canonical class of sensory complex for C4-dicarboxylates.
Sci Rep, 7, 2017
|
|
5LWL
 
 | | MaeR D54A mutant response regulator bound to sulfate | | Descriptor: | SULFATE ION, Transcriptional regulatory protein | | Authors: | Miguel-Romero, L, Casino, P, Landete, J.M, Monedero, V, Zuniga, M, Marina, A. | | Deposit date: | 2016-09-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The malate sensing two-component system MaeKR is a non-canonical class of sensory complex for C4-dicarboxylates.
Sci Rep, 7, 2017
|
|
4KP4
 
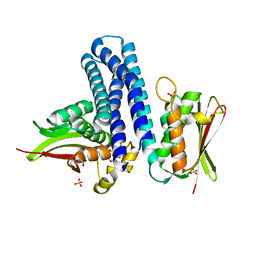 | | Deciphering cis-trans directionality and visualizing autophosphorylation in histidine kinases. | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Osmolarity sensor protein EnvZ, ... | | Authors: | Casino, P, Miguel-Romero, L, Marina, A. | | Deposit date: | 2013-05-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing autophosphorylation in histidine kinases.
Nat Commun, 5, 2014
|
|
5O8Z
 
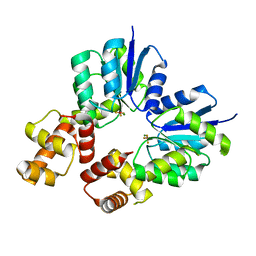 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
5O8Y
 
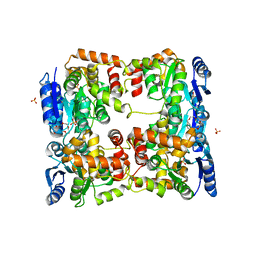 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
6PAJ
 
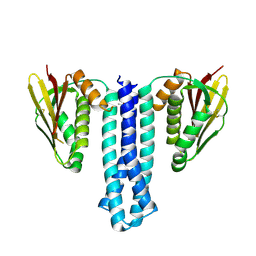 | |
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5I04
 
 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
6EO2
 
 | |
6EO3
 
 | |
6RGZ
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH8
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RFV
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH2
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH7
 
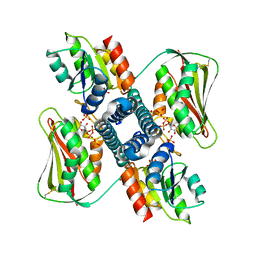 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH1
 
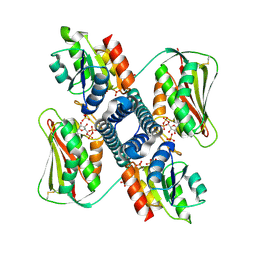 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH0
 
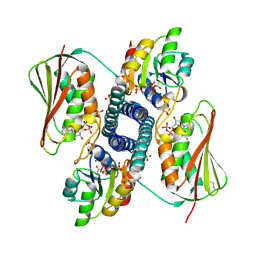 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
