7QFU
 
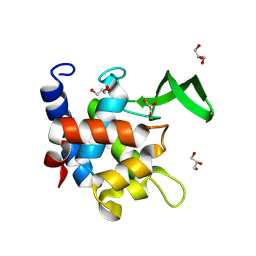 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
2MKX
 
 | |
8P8E
 
 | |
6SMK
 
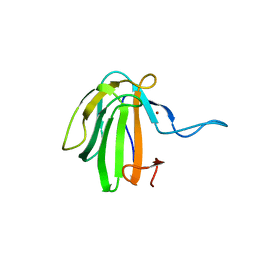 | | Crystal structure of catalytic domain A109H mutant of prophage-encoded M23 protein EnpA from Enterococcus faecalis. | | Descriptor: | Peptidase_M23 domain-containing protein, ZINC ION | | Authors: | Malecki, P.H, Mitkowski, P, Czapinska, H, Sabala, I. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Characterization of EnpA D,L-Endopeptidase from Enterococcus faecalis Prophage Provides Insights into Substrate Specificity of M23 Peptidases.
Int J Mol Sci, 22, 2021
|
|
6RK4
 
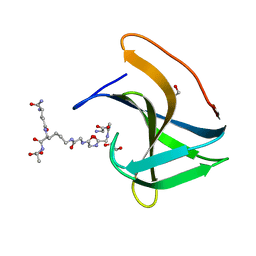 | | Lysostaphin SH3b P4-G5 complex, synchrotron dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, 1,2-ETHANEDIOL, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
6RJE
 
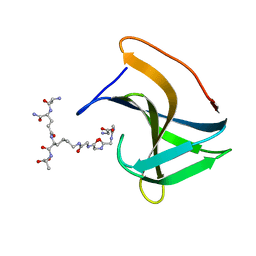 | | Lysostaphin SH3b P4-G5 complex, homesource dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
1XE4
 
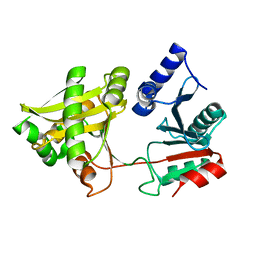 | | Crystal Structure of Weissella viridescens FemX (K36M) Mutant | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Arthur, M, Mayer, C. | | Deposit date: | 2004-09-09 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of the UDP-MurNAc-Pentapeptide-Binding Cavity of the FemX Alanyl Transferase from Weissella viridescens
J.Bacteriol., 187, 2005
|
|
1XF8
 
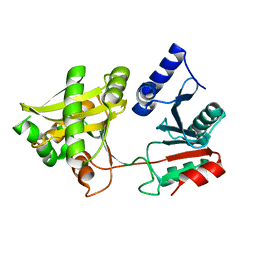 | | Crystal Structure of Weissella viridescens FemX (Y254F) Mutant | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Arthur, M, Mayer, C. | | Deposit date: | 2004-09-14 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of the UDP-MurNAc-Pentapeptide-Binding Cavity of the FemX Alanyl Transferase from Weissella viridescens
J.BACTERIOL., 187, 2005
|
|
1XIX
 
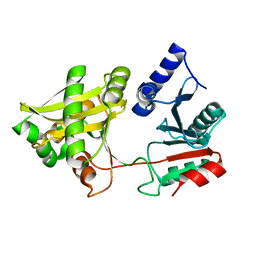 | |
4PI9
 
 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4PI7
 
 | | Crystal structure of S. Aureus Autolysin E in complex with disaccharide NAM-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PIA
 
 | | Crystal structure of S. Aureus Autolysin E | | Descriptor: | Autolysin E, CHLORIDE ION | | Authors: | Mihelic, M, Renko, M, Dobersek, A, Bedrac, L, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4PI8
 
 | | Crystal structure of catalytic mutant E138A of S. Aureus Autolysin E in complex with disaccharide NAG-NAM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
