5FO3
 
 | | ZapC cell division regulator from E. coli | | Descriptor: | CELL DIVISION PROTEIN ZAPC | | Authors: | Ortiz, C, Kureisaite-Ciziene, D, Schmitz, F, Vicente, M, Lowe, J. | | Deposit date: | 2015-11-18 | | Release date: | 2015-11-25 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Z-Ring Associated Cell Division Protein Zapc from Escherichia Coli.
FEBS Lett., 589, 2015
|
|
5LY3
 
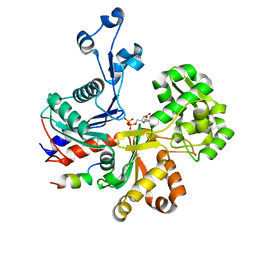 | |
5LY5
 
 | | Arcadin-1 from Pyrobaculum calidifontis | | Descriptor: | arcadin-1 | | Authors: | Izore, T, Lowe, J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crenactin forms actin-like double helical filaments regulated by arcadin-2.
Elife, 5, 2016
|
|
5MN4
 
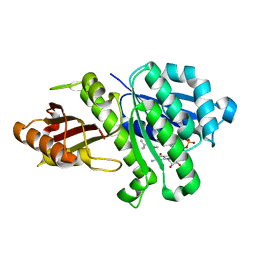 | | S. aureus FtsZ 12-316 F138A GDP Open form (1FOf) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN8
 
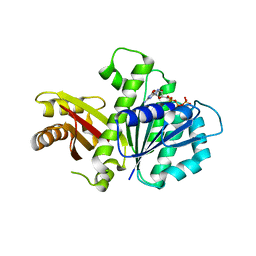 | | S. aureus FtsZ 12-316 F138A GTP Closed form (5FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MW1
 
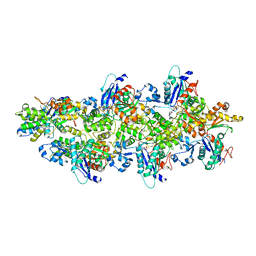 | |
5MN5
 
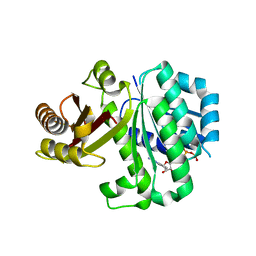 | | S. aureus FtsZ 12-316 T66W GTP Closed form (2TCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN6
 
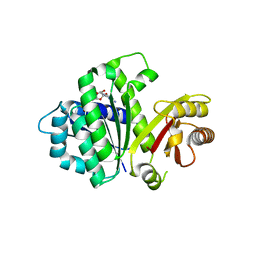 | | S. aureus FtsZ 12-316 F138A GDP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5MN7
 
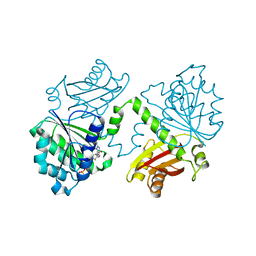 | | S. aureus FtsZ 12-316 F138A GTP Closed form (3FCm) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
5N8P
 
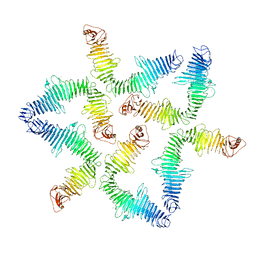 | |
4A2B
 
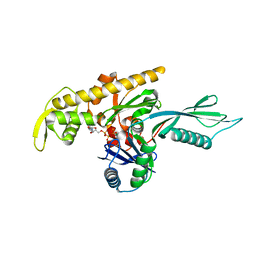 | |
4A2A
 
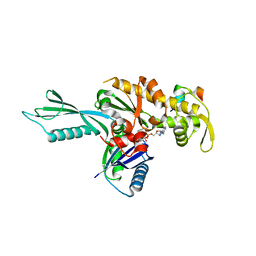 | | Thermotoga maritima FtsA:FtsZ(336-351) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN FTSA, PUTATIVE, ... | | Authors: | Szwedziak, P, Lowe, J. | | Deposit date: | 2011-09-23 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ftsa Forms Actin-Like Protofilaments
Embo J., 31, 2012
|
|
2WD5
 
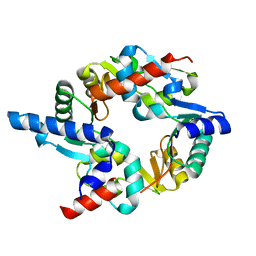 | | SMC hinge heterodimer (Mouse) | | Descriptor: | STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 1A, STRUCTURAL MAINTENANCE OF CHROMOSOMES PROTEIN 3 | | Authors: | Michie, K.A, Haering, C.H, Nasmyth, K, Lowe, J. | | Deposit date: | 2009-03-20 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Positively Charged Channel within the Smc1/Smc3 Hinge Required for Sister Chromatid Cohesion.
Embo J., 30, 2011
|
|
2IUU
 
 | | P. aeruginosa FtsK motor domain, hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA TRANSLOCASE FTSK | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
2IUT
 
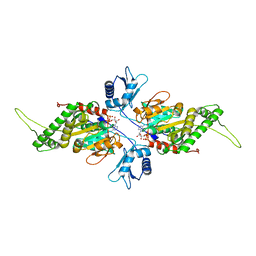 | | P. aeruginosa FtsK motor domain, dimeric | | Descriptor: | DNA TRANSLOCASE FTSK, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
2IUS
 
 | | E. coli FtsK motor domain | | Descriptor: | DNA TRANSLOCASE FTSK | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
2JEE
 
 | | Xray structure of E. coli YiiU | | Descriptor: | CELL DIVISION PROTEIN ZAPB | | Authors: | Moller-Jensen, J, Gerdes, K, Lowe, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Coiled-Coil Cell Division Factor Zapb Stimulates Z Ring Assembly and Cell Division.
Mol.Microbiol., 68, 2008
|
|
1MWM
 
 | | ParM from plasmid R1 ADP form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM | | Authors: | Van den Ent, F, Moller-Jensen, J, Amos, L.A, Gerdes, K, Lowe, J. | | Deposit date: | 2002-09-30 | | Release date: | 2003-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | F-actin-like filaments formed by plasmid segregation protein ParM
EMBO J., 21, 2002
|
|
1MWK
 
 | | ParM from plasmid R1 APO form | | Descriptor: | ParM | | Authors: | Van den Ent, F, Moller-Jensen, J, Amos, L.A, Gerdes, K, Lowe, J. | | Deposit date: | 2002-09-30 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | F-actin-like filaments formed by plasmid segregation protein ParM
EMBO J., 21, 2002
|
|
1HJZ
 
 | | Crystal structure of AF1521 protein containing a macroH2A domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL PROTEIN AF1521 | | Authors: | Allen, M.D, Buckle, A.M, Cordell, S.C, Lowe, J, Bycroft, M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Af1521 a Protein from Archaeoglobus Fulgidus with Homology to the Non-Histone Domain of Macroh2A
J.Mol.Biol., 330, 2003
|
|
4ASS
 
 | |
4ASO
 
 | |
4B46
 
 | |
1QHL
 
 | |
2XJ4
 
 | |
