2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
1ZL8
 
 | | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | | Descriptor: | LIN-7, Peripheral plasma membrane protein CASK | | Authors: | Petrosky, K.Y, Ou, H.D, Lohr, F, Dotsch, V, Lim, W.A. | | Deposit date: | 2005-05-05 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A General Model for Preferential Hetero-oligomerization of LIN-2/7 Domains: Mechanism Underlying Directed Assembly of Supramolecular Signaling Complexes
J.Biol.Chem., 280, 2005
|
|
5N6R
 
 | | Solution structure of the Dbl-homology domain of Bcr-Abl | | Descriptor: | Breakpoint cluster region protein | | Authors: | Reckel, S, Lohr, F, Buchner, L, Guntert, P, Dotsch, V, Hantschel, O. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
1L6U
 
 | | NMR STRUCTURE OF OXIDIZED ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1L6V
 
 | | STRUCTURE OF REDUCED BOVINE ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
2AYX
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYY
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
6FGS
 
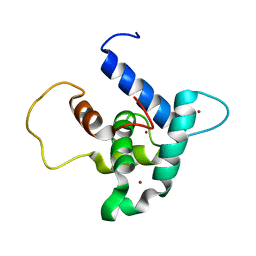 | | Solution structure of p300Taz2-p73TA1 | | Descriptor: | Histone acetyltransferase p300,Tumor protein p73, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
6FGN
 
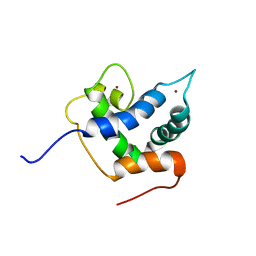 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
2K2Q
 
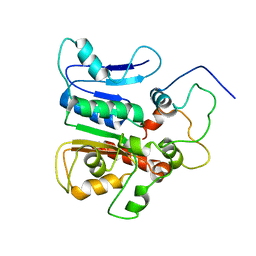 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
2K0L
 
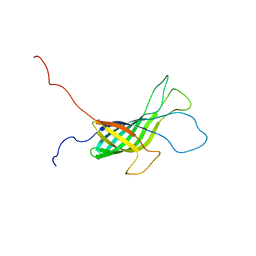 | | NMR structure of the transmembrane domain of the Outer Membrane Protein A from Klebsiella pneumoniae in DHPC micelles. | | Descriptor: | Outer membrane protein A | | Authors: | Renault, M, Saurel, O, Gervais, V, Lohr, F, Reat, V, Piotto, M, Milon, A. | | Deposit date: | 2008-02-04 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution state NMR structure and dynamics of KpOmpA, a 210 residue transmembrane domain possessing a high potential for immunological applications.
J.Mol.Biol., 385, 2009
|
|
2JO7
 
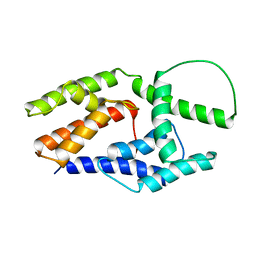 | | Solution structure of the adhesion protein Bd37 from Babesia divergens | | Descriptor: | Glycosylphosphatidylinositol-anchored merozoite surface protein | | Authors: | Auguin, D, Yang, Y, Lohr, F, Arold, S, Schetters, T, Precigout, E, Gorenflot, A, Delbecq, S, Roumestand, C. | | Deposit date: | 2007-02-26 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the Adhesion Protein Bd37 from Babesia divergens Reveals Structural Homology with Eukaryotic Proteins Involved in Membrane Trafficking
J.Mol.Biol., 375, 2007
|
|
2GVX
 
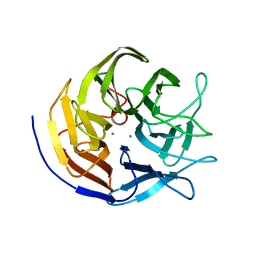 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
1DF3
 
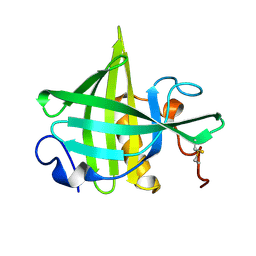 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
8PH4
 
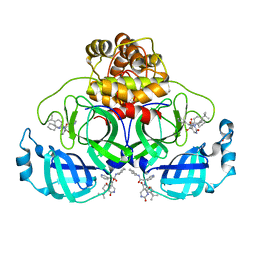 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
8CHP
 
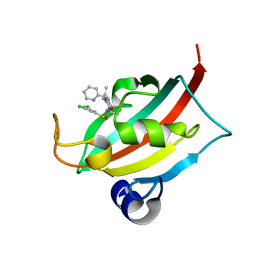 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHR
 
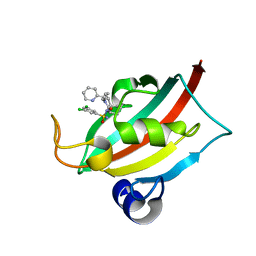 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((R)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHK
 
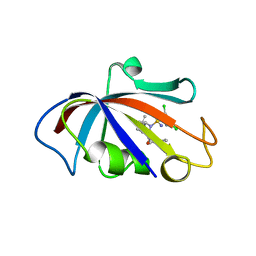 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHI
 
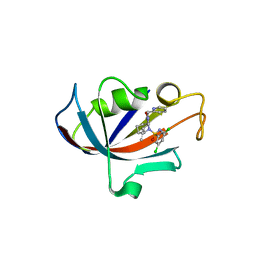 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHQ
 
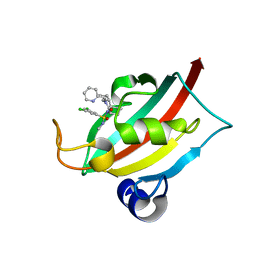 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHL
 
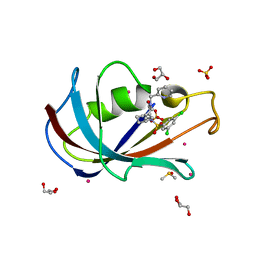 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHN
 
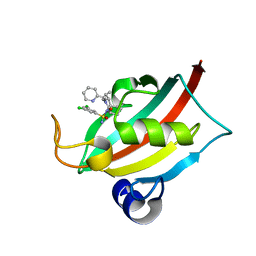 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHM
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
