1DF3
 
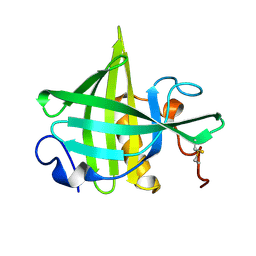 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
2GDY
 
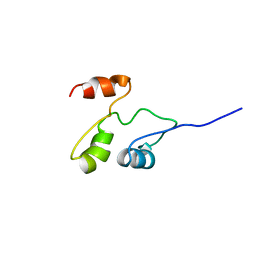 | | Solution structure of the B. brevis TycC3-PCP in A-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2GDX
 
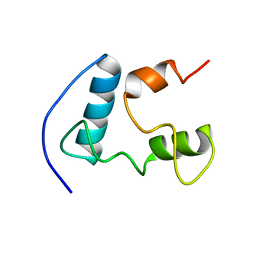 | | Solution structure of the B. brevis TycC3-PCP in H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
1QXN
 
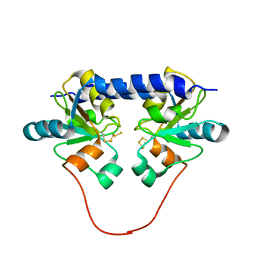 | | Solution Structure of the 30 kDa Polysulfide-sulfur Transferase Homodimer from Wolinella Succinogenes | | Descriptor: | PENTASULFIDE-SULFUR, sulfide dehydrogenase | | Authors: | Lin, Y.J, Dancea, F, Loehr, F, Klimmek, O, Pfeiffer-Marek, S, Nilges, M, Wienk, H, Kroeger, A, Rueterjans, H. | | Deposit date: | 2003-09-08 | | Release date: | 2004-02-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 30 kDa Polysulfide-Sulfur Transferase Homodimer from Wolinella succinogenes
Biochemistry, 43, 2004
|
|
1SR2
 
 | |
2FWL
 
 | | The cytochrome c552/CuA complex from Thermus thermophilus | | Descriptor: | Cytochrome c oxidase subunit II, Cytochrome c-552, DINUCLEAR COPPER ION, ... | | Authors: | Muresanu, L, Pristovsek, P, Loehr, F, Maneg, O, Mukrasch, M.D, Rueterjans, H, Ludwig, B, Luecke, C. | | Deposit date: | 2006-02-02 | | Release date: | 2006-03-28 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The electron transfer complex between cytochrome c552 and the CuA domain of the Thermus thermophilus ba3 oxidase - a combined NMR and computational approach
J.Biol.Chem., 281, 2006
|
|
2GDW
 
 | | Solution structure of the B. brevis TycC3-PCP in A/H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
2IAX
 
 | | Crystal structure of squid ganglion DFPase D232S mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
2IAW
 
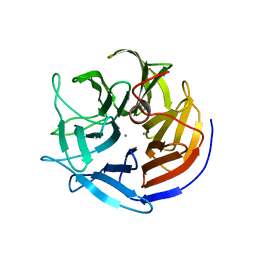 | | Crystal structure of squid ganglion DFPase N175D mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
2IAV
 
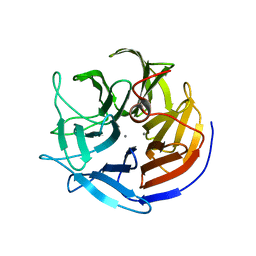 | | Crystal structure of squid ganglion DFPase H287A mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
2LO6
 
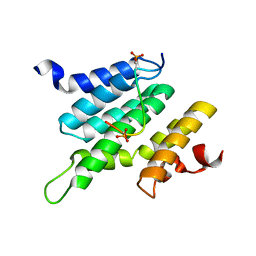 | | Structure of Nrd1 CID bound to phosphorylated RNAP II CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Protein NRD1 | | Authors: | Kubicek, K, Cerna, H, Pasulka, J, Holub, P, Hrossova, D, Loehr, F, Hofr, C, Vanacova, S, Stefl, R. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1.
Genes Dev., 26, 2012
|
|
5HKH
 
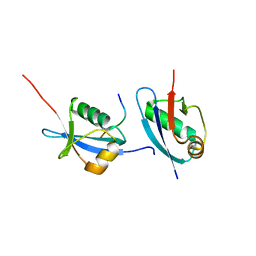 | | Crystal structure of Ufm1 in complex with UBA5 | | Descriptor: | ASP-ASN-GLU-TRP-GLY-ILE-GLU-LEU-VAL, Ubiquitin-fold modifier 1 | | Authors: | Huber, J, Doetsch, V, Rogov, V.V, Akutsu, M. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation.
J.Biol.Chem., 291, 2016
|
|
7OVC
 
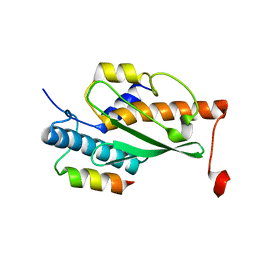 | | Structure of the human UFC1 protein in complex with the UBA5 C-terminal UFC1-binding motif. | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Wesch, W, Loehr, F, Rogova, N, Doetsch, V, Rogov, V.V. | | Deposit date: | 2021-06-14 | | Release date: | 2021-08-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Concerted Action of UBA5 C-Terminal Unstructured Regions Is Important for Transfer of Activated UFM1 to UFC1.
Int J Mol Sci, 22, 2021
|
|
4MRT
 
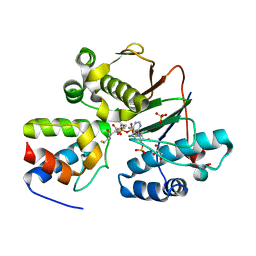 | | Structure of the Phosphopantetheine Transferase Sfp in Complex with Coenzyme A and a Peptidyl Carrier Protein | | Descriptor: | 4'-phosphopantetheinyl transferase sfp, COENZYME A, GLYCEROL, ... | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
6H8C
 
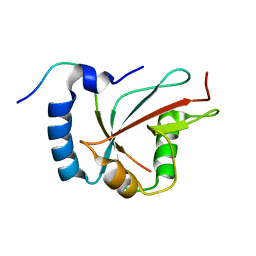 | | Structure of the human GABARAPL2 protein in complex with the UBA5 LIR motif | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 2, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Huber, J, Loehr, F, Gruber, J, Akutsu, M, Guentert, P, Doetsch, V, Rogov, V.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Autophagy, 16, 2020
|
|
1I6D
 
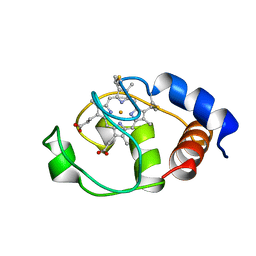 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
2RP5
 
 | |
2RP4
 
 | |
1P4W
 
 | | Solution structure of the DNA-binding domain of the Erwinia amylovora RcsB protein | | Descriptor: | rcsB | | Authors: | Pristovsek, P, Sengupta, K, Loehr, F, Schaefer, B, Wehland von Trebra, M, Rueterjans, H, Bernhard, F. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box.
J.Biol.Chem., 278, 2003
|
|
2MYX
 
 | | Structure of the CUE domain of yeast Cue1 | | Descriptor: | Coupling of ubiquitin conjugation to ER degradation protein 1 | | Authors: | Kniss, A, Rogov, V.V, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2015-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.
Mol.Cell, 62, 2016
|
|
2MW4
 
 | | Tetramerization domain of the Ciona intestinalis p53/p73-b transcription factor protein | | Descriptor: | Transcription factor protein | | Authors: | Heering, J.P, Jonker, H.R.A, Loehr, F, Schwalbe, H, Doetsch, V. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain.
Protein Sci., 25, 2016
|
|
1I6E
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1PM6
 
 | | Solution Structure of Full-Length Excisionase (Xis) from Bacteriophage HK022 | | Descriptor: | Excisionase | | Authors: | Rogov, V.V, Luecke, C, Muresanu, L, Wienk, H, Kleinhaus, I, Werner, K, Loehr, F, Pristovsek, P, Rueterjans, H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the full-length excisionase from bacteriophage HK022.
Eur.J.Biochem., 270, 2003
|
|
2KX7
 
 | | Solution structure of the E.coli RcsD-ABL domain (residues 688-795) | | Descriptor: | Sensor-like histidine kinase yojN | | Authors: | Rogov, V.V, Schmoee, K, Rogova, N.Y, Loehr, F, Bernhard, F, Doetsch, V. | | Deposit date: | 2010-04-27 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Rcs Phosphotransfer: The Newly Identified RcsD-ABL Domain Enhances Interaction with the Response Regulator RcsB.
Structure, 19, 2011
|
|
2L6X
 
 | | Solution NMR Structure of Proteorhodopsin. | | Descriptor: | Green-light absorbing proteorhodopsin, RETINAL | | Authors: | Reckel, S, Gottstein, D, Stehle, J, Loehr, F, Takeda, M, Silvers, R, Kainosho, M, Glaubitz, C, Bernhard, F, Schwalbe, H, Guntert, P, Doetsch, V, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-09 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of proteorhodopsin.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
