3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
6M1K
 
 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
8HM1
 
 | |
8HM2
 
 | |
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM3
 
 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSJ
 
 | | Crystal structure of 1,6-APD (2-ANIMOPHENOL-1,6-DIOXYGENASE) complexed with intermediate products | | Descriptor: | (2Z,4Z)-2-imino-6-oxohex-4-enoic acid, (3E)-3-iminooxepin-2(3H)-one, 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSH
 
 | | Crystal structure of native 1,6-APD (with Iron), 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, FE (II) ION | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6SMF
 
 | |
6SME
 
 | |
7CCF
 
 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | Descriptor: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GWE
 
 | | cytochrome P450 CREJ | | Descriptor: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Dong, S, liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-09-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3BM8
 
 | |
5XUA
 
 | |
5XUB
 
 | |
4DGX
 
 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
4DGP
 
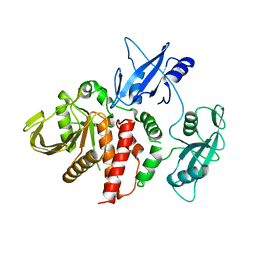 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
7DQ9
 
 | |
6ITS
 
 | |
6HSR
 
 | |
6HSQ
 
 | |
