1BKJ
 
 | | NADPH:FMN OXIDOREDUCTASE FROM VIBRIO HARVEYI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH-FLAVIN OXIDOREDUCTASE, PHOSPHATE ION | | Authors: | Tanner, J.J, Lei, B, TU, S.-C, Krause, K.L. | | Deposit date: | 1998-07-08 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin reductase P: structure of a dimeric enzyme that reduces flavin.
Biochemistry, 35, 1996
|
|
2MOQ
 
 | | Solution Structure and Molecular determinants of Hemoglobin Binding of the first NEAT Domain of IsdB in Staphylococcus aureus | | Descriptor: | Iron-regulated surface determinant protein B | | Authors: | Fonner, B.A, Tripet, B.P, Eilers, B.J, Stanisich, J, Sullivan-Springhetti, R.K, Moore, R, Lui, M, Lei, B, Copie, V. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Molecular Determinants of Hemoglobin Binding of the First NEAT Domain of IsdB in Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
6Q3P
 
 | | Atomic resolution crystal structure of an AAB collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading Chain of the AAB Collagen Heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
6Q41
 
 | | Atomic resolution crystal structure of a BAA collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading chain of the BAA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
1BLJ
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
2WUH
 
 | | Crystal structure of the DDR2 discoidin domain bound to a triple- helical collagen peptide | | Descriptor: | COLLAGEN PEPTIDE, DISCOIDIN DOMAIN RECEPTOR 2 | | Authors: | Carafoli, F, Bihan, D, Stathopoulos, S, Konitsiotis, A.D, Kvansakul, M, Farndale, R.W, Leitinger, B, Hohenester, E. | | Deposit date: | 2009-10-05 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Insight Into Collagen Recognition by Discoidin Domain Receptor 2
Structure, 17, 2009
|
|
2LFB
 
 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
6Q43
 
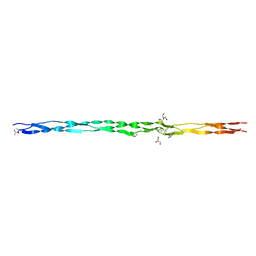 | | Atomic resolution crystal structure of an ABA collagen heterotrimer | | Descriptor: | GLYCEROL, Leading Chain of the ABA collagen heterotrimer, Middle Chain of the ABA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
6Y23
 
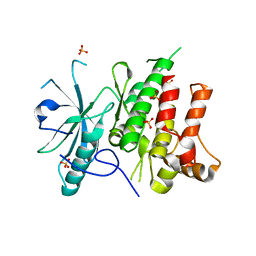 | |
7S3F
 
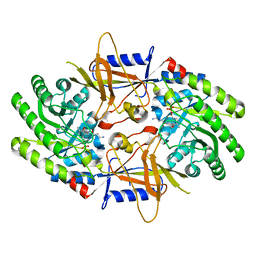 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
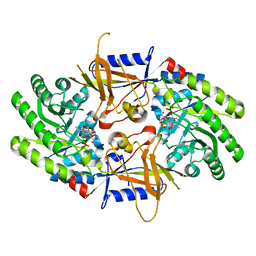 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
1X70
 
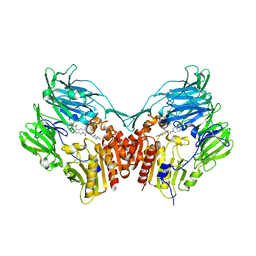 | | HUMAN DIPEPTIDYL PEPTIDASE IV IN COMPLEX WITH A BETA AMINO ACID INHIBITOR | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, D, Wang, L, Beconi, M, Eiermann, G.J, Fisher, M.H, He, H, Hickey, G.J, Leiting, B, Lyons, K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (2R)-4-Oxo-4-[3-(Trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin- 7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: A Potent, Orally Active Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes
J.Med.Chem., 48, 2005
|
|
4AG4
 
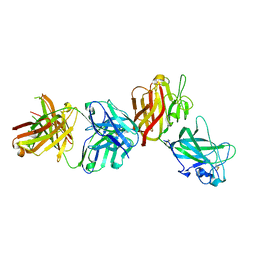 | | Crystal structure of a DDR1-Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1, ... | | Authors: | Carafoli, F, Mayer, M.C, Shiraishi, K, Pecheva, M.A, Chan, L.Y, Nan, R, Leitinger, B, Hohenester, E. | | Deposit date: | 2012-01-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Discoidin Domain Receptor 1 Extracellular Region Bound to an Inhibitory Fab Fragment Reveals Features Important for Signaling.
Structure, 20, 2012
|
|
1N10
 
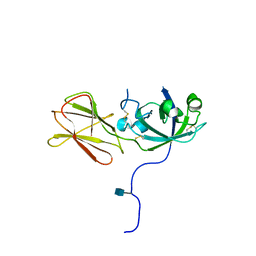 | | Crystal Structure of Phl p 1, a Major Timothy Grass Pollen Allergen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pollen allergen Phl p 1 | | Authors: | Fedorov, A.A, Ball, T, Leistler, B, Valenta, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-10-16 | | Release date: | 2003-01-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystal Structure of Phl p 1, a Major Timothy Grass Pollen Allergen
To be Published
|
|
1BLK
 
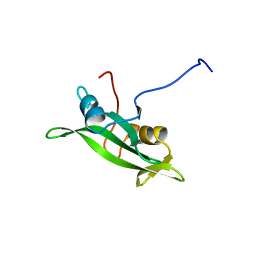 | | NMR ENSEMBLE OF BLK SH2 DOMAIN USING CHEMICAL SHIFT REFINEMENT, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
2LWB
 
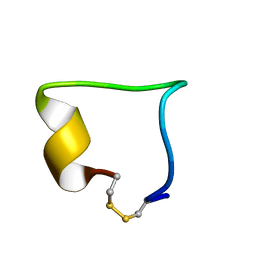 | | Structural model of BAD-1 repeat loop by NMR | | Descriptor: | Adhesin WI-1 | | Authors: | Brandhorst, T, Klein, B, Tonelli, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a fungal adhesin that mimics thrombospondin-1 by binding heparin sulfate glycosaminoglycan and suppressing T cell activation via interaction with CD47
To be Published
|
|
2BKJ
 
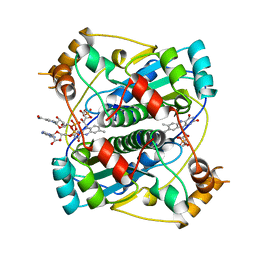 | |
2LHR
 
 | |
2Q7A
 
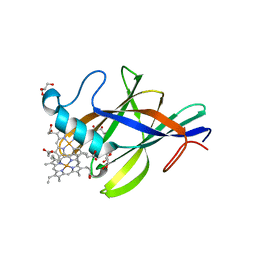 | | Crystal structure of the cell surface heme transfer protein Shp | | Descriptor: | Cell surface heme-binding protein, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Aranda IV, R, Worley, C.E, Bitto, E, Phillips Jr, G.N. | | Deposit date: | 2007-06-06 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp.
J.Mol.Biol., 374, 2007
|
|
2FJP
 
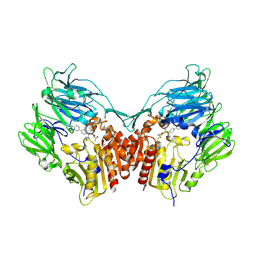 | | Human dipeptidyl peptidase IV/CD26 in complex with an inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-{(1S,2S)-2-AMINO-1-[(DIMETHYLAMINO)CARBONYL]-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-3-OXOPROPYL}PHENYL)-1H-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-4-IUM, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W. | | Deposit date: | 2006-01-03 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (2S,3S)-3-Amino-4-(3,3-difluoropyrrolidin-1-yl)-N,N-dimethyl-4-oxo-2-(4-[1,2,4]triazolo[1,5-a]- pyridin-6-ylphenyl)butanamide: a selective alpha-amino amide dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
J.Med.Chem., 49, 2006
|
|
3C43
 
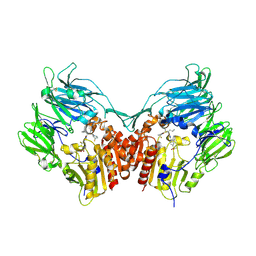 | | Human dipeptidyl peptidase IV/CD26 in complex with a flouroolefin inhibitor | | Descriptor: | (2S,3S)-4-cyclopropyl-3-{(3R,5R)-3-[2-fluoro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazolidin-5-yl}-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxobutan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Edmondson, S.D, Weber, A.E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fluoroolefins as amide bond mimics in dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2QOE
 
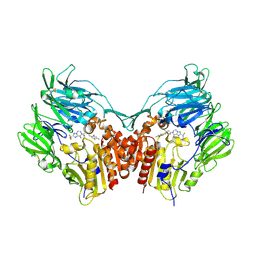 | | Human Dipeptidyl Peptidase IV in complex with a Triazolopiperazine-based beta amino acid Inhibitor | | Descriptor: | (2R)-4-[(8R)-8-METHYL-2-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRAZIN-7(8H)-YL]-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-07-20 | | Release date: | 2007-11-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis, and biological evaluation of triazolopiperazine-based beta-amino amides as potent, orally active dipeptidyl peptidase IV (DPP-4) inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QT9
 
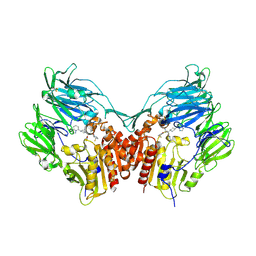 | | Human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl cyclohexylalanine inhibitor | | Descriptor: | (2S,3S)-3-AMINO-4-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-N,N-DIMETHYL-4-OXO-2-(TRANS-4-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-5-YLCYCLOH EXYL)BUTANAMIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 4-Arylcyclohexylalanine analogs as potent, selective, and orally active inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6T28
 
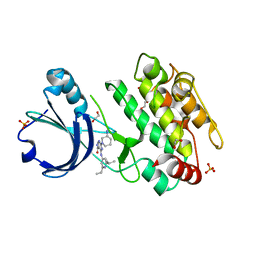 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
