8IWL
 
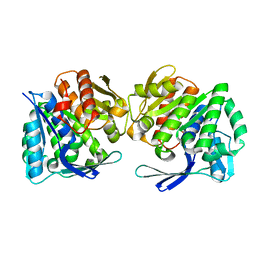 | | A.baumannii Uncharacterized sugar kinase ydjH | | Descriptor: | Uncharacterized sugar kinase YdjH | | Authors: | Lee, G.H, Park, H.H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of YdjH from Acinetobacter baumannii revealed an active site of YdjH family sugar kinase.
Biochem.Biophys.Res.Commun., 664, 2023
|
|
7WZM
 
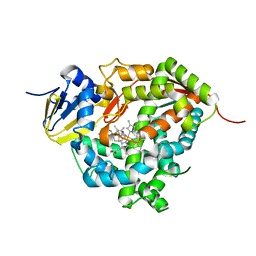 | | Crystal structure of Cytochrome P450 184A1 from streptomyces avermitilis in complex with Oleic acid | | Descriptor: | OLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Kim, V.C, Kim, D.G, Lee, S.G, Lee, G.H, Lee, S.A, Kang, L.W. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Cytochrome P450 184A1 from streptomyces avermitilis in complex with Oleic acid
To Be Published
|
|
7WZL
 
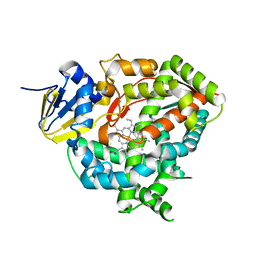 | | Crystal structure of Cytochrome P450 184A1 from streptomyces avermitilis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Kim, V.C, Kim, D.G, Lee, S.G, Lee, G.H, Lee, S.A, Kang, L.W. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cytochrome P450 184A1 from streptomyces avermitilis
To Be Published
|
|
5XEW
 
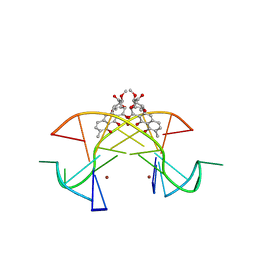 | | Crystal structure of the [Ni2+-(chromomycin A3)2]-CCG repeats complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Tseng, W.H, Wu, P.C, Hou, M.H. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel-Chromomycin Complex Resulting in Large-Scale DNA Deformation
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8JQZ
 
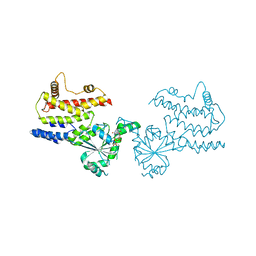 | | Crystal Structure of GppNHp-bound mIRGB10 | | Descriptor: | Immunity-related GTPase family member b10, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ha, H.J, Park, H.H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of IRGB10 oligomerization by GTP hydrolysis.
Front Immunol, 14, 2023
|
|
8JQY
 
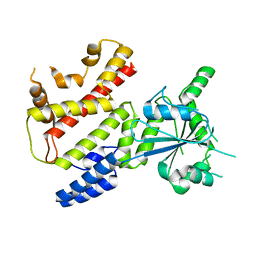 | |
8K2Y
 
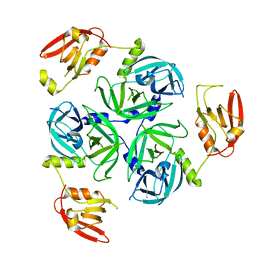 | | Crystal structure of MucD | | Descriptor: | serine endoprotease DegP-like protein MucD | | Authors: | Kim, J.H, Park, H.H. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of MucD from Pseudomonas syringae revealed N-terminal loop-mediated trimerization of HtrA-like serine protease.
Biochem.Biophys.Res.Commun., 688, 2023
|
|
5Z70
 
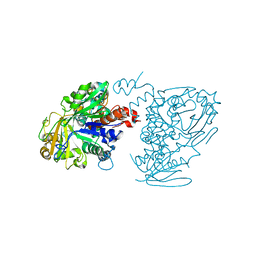 | |
8HJJ
 
 | | Anti-CRISPR protein AcrIC9 | | Descriptor: | Anti-CRISPR protein Type I-C9 | | Authors: | Kang, Y.J, Park, H.H. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of AcrIC9 revealing the putative inhibitory mechanism of AcrIC9 against the type IC CRISPR-Cas system.
Iucrj, 10, 2023
|
|
