2Y31
 
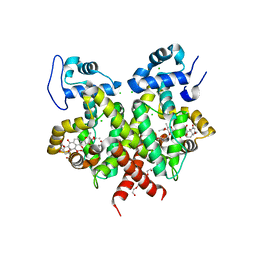 | | Simocyclinone C4 bound form of TetR-like repressor SimR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
2Y30
 
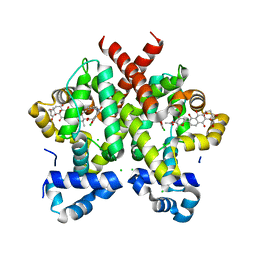 | | Simocyclinone D8 bound form of TetR-like repressor SimR | | Descriptor: | CHLORIDE ION, PUTATIVE REPRESSOR SIMREG2, SIMOCYCLINONE D8 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
2Y2Z
 
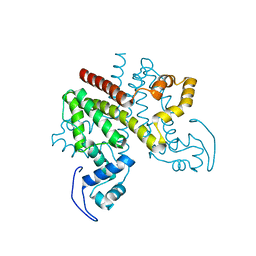 | | ligand-free form of TetR-like repressor SimR | | Descriptor: | PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
3ZQL
 
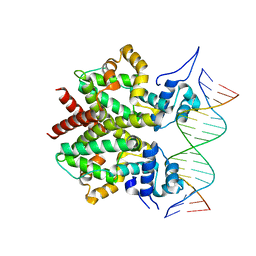 | | DNA-bound form of TetR-like repressor SimR | | Descriptor: | 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*CP*CP*GP*TP*AP*DCP *GP*AP*A)-3', 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*GP*CP*GP*TP*AP*DCP *GP*AP*A)-3', PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Schumacher, M.A, Lawson, D.M, Brennan, R.G, Buttner, M.J. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Crystal Structure of the Tetr Family Transcriptional Repressor Simr Bound to DNA and the Role of a Flexible N-Terminal Extension in Minor Groove Binding.
Nucleic Acids Res., 39, 2011
|
|
7OL9
 
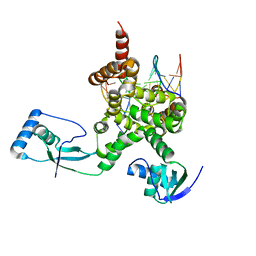 | |
6Y93
 
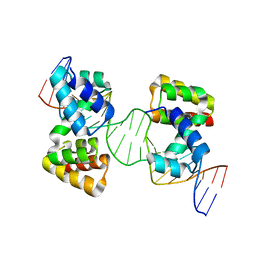 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
2Y3P
 
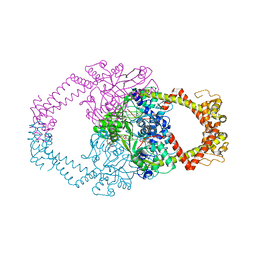 | | Crystal structure of N-terminal domain of GyrA with the antibiotic simocyclinone D8 | | Descriptor: | DNA GYRASE SUBUNIT A, MAGNESIUM ION, SIMOCYCLINONE D8 | | Authors: | Edwards, M.J, Flatman, R.H, Mitchenall, L.A, Stevenson, C.E.M, Le, T.B.K, Clarke, T.A, McKay, A.R, Fiedler, H.-P, Buttner, M.J, Lawson, D.M, Maxwell, A. | | Deposit date: | 2010-12-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8, Bound to DNA Gyrase.
Science, 326, 2009
|
|
6S6H
 
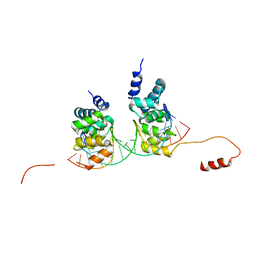 | | Crystal structure of the DNA binding domain of the chromosome-partitioning protein ParB complexed to the centromeric parS site | | Descriptor: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*C)-3'), GLYCEROL | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Tan, E.X, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2019-07-03 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
7NFU
 
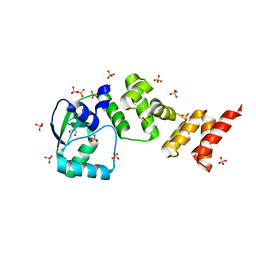 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
7NG0
 
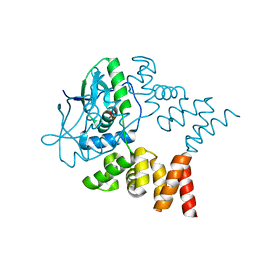 | | Crystal structure of N- and C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
8QA9
 
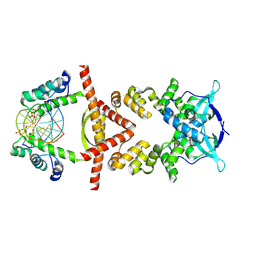 | | Crystal structure of the RK2 plasmid encoded co-complex of the C-terminally truncated transcriptional repressor protein KorB complexed with the partner repressor protein KorA bound to OA-DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*A)-3'), SULFATE ION, Transcriptional repressor protein KorB, ... | | Authors: | McLean, T.C, Mundy, J.E.A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2023-08-22 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the RK2 plasmid encoded co-complex of the C-terminally truncated transcriptional repressor protein KorB complexed with the partner repressor protein KorA bound to OA-DNA
To Be Published
|
|
8QA8
 
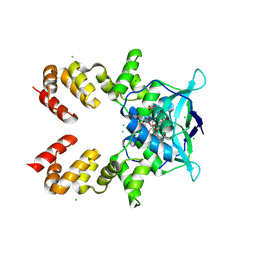 | |
6AMK
 
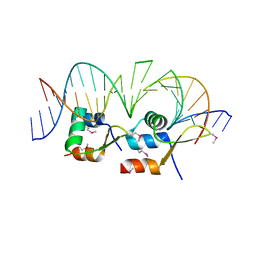 | | Structure of Streptomyces venezuelae BldC-whiI opt complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*TP*TP*AP*CP*CP*CP*GP*AP*AP*TP*TP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*AP*TP*TP*CP*GP*GP*GP*TP*AP*AP*TP*TP*CP*GP*GP*GP*CP*A)-3'), Putative DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development.
Nat Commun, 9, 2018
|
|
6AMA
 
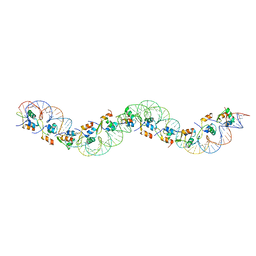 | |
7BM8
 
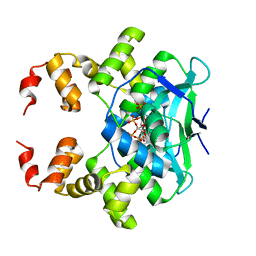 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed with CTP-gamma-S | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Chromosome-partitioning protein ParB, MAGNESIUM ION | | Authors: | Jalal, A.S, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
3ZPL
 
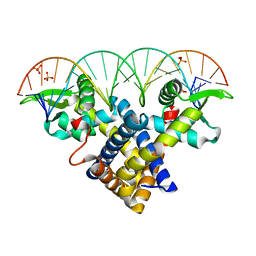 | | Crystal structure of Sco3205, a MarR family transcriptional regulator from Streptomyces coelicolor, in complex with DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*TP*GP*AP*GP*AP*TP*CP*TP *CP*AP*AP*TP*CP*TP*TP*DT)-3', PHOSPHATE ION, PUTATIVE MARR-FAMILY TRANSCRIPTIONAL REPRESSOR | | Authors: | Stevenson, C.E.M, Assaad, A, Lawson, D.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of DNA Sequence Recognition by a Streptomycete Marr Family Transcriptional Regulator Through Surface Plasmon Resonance and X-Ray Crystallography.
Nucleic Acids Res., 41, 2013
|
|
6T1F
 
 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed to the centromeric parS site | | Descriptor: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*CP*C)-3') | | Authors: | Jalal, A.S.B, Pastrana, C.L, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Moreno-Herrero, F, Le, T.B.K. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
