7VUF
 
 | |
7VUK
 
 | |
1IYI
 
 | | Crystal structure of hematopoietic prostaglandin D synthase | | Descriptor: | CALCIUM ION, GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE | | Authors: | Inoue, T. | | Deposit date: | 2002-08-26 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of metal activation of human hematopoietic prostaglandin D synthase
NAT.STRUCT.BIOL., 10, 2003
|
|
1IYH
 
 | | Crystal structure of hematopoietic prostaglandin D synthase | | Descriptor: | GLUTATHIONE, HEMATOPOIETIC PROSTAGLANDIN D SYNTHASE, MAGNESIUM ION | | Authors: | Inoue, T. | | Deposit date: | 2002-08-26 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of metal activation of human hematopoietic prostaglandin D synthase
NAT.STRUCT.BIOL., 10, 2003
|
|
5H71
 
 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate trisaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgQ2, CALCIUM ION, ... | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
5H6U
 
 | | Structure of alginate-binding protein AlgQ2 in complex with an alginate pentasaccharide | | Descriptor: | AlgQ2, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Uenishi, K, Kaneko, A, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2016-11-15 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate
J. Biol. Chem., 292, 2017
|
|
6KV0
 
 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LK1
 
 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
1EWV
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1EWK
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1ZAB
 
 | | Crystal Structure of Mouse Cytidine Deaminase Complexed with 3-Deazauridine | | Descriptor: | 1-((2R,3R,4S,5R)-TETRAHYDRO-3,4-DIHYDROXY-5-(HYDROXYMETHYL)FURAN-2-YL)PYRIDINE-2,4(1H,3H)-DIONE, Cytidine deaminase, SULFATE ION, ... | | Authors: | Teh, A.H. | | Deposit date: | 2005-04-06 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The 1.48 A Resolution Crystal Structure of the Homotetrameric Cytidine Deaminase from Mouse
Biochemistry, 45, 2006
|
|
2FR6
 
 | | Crystal Structure of Mouse Cytidine Deaminase Complexed with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, AMMONIA, Cytidine deaminase, ... | | Authors: | Teh, A.H. | | Deposit date: | 2006-01-19 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The 1.48 A Resolution Crystal Structure of the Homotetrameric Cytidine Deaminase from Mouse
Biochemistry, 45, 2006
|
|
2FR5
 
 | |
5X9Y
 
 | |
5Z41
 
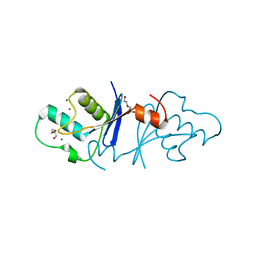 | | Aquifex aeolicus MutL endonuclease domain with a single zinc ion. | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, MAGNESIUM ION, ... | | Authors: | Fukui, K, Yano, T. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Multiple zinc ions maintain the open conformation of the catalytic site in the DNA mismatch repair endonuclease MutL from Aquifex aeolicus
FEBS Lett., 592, 2018
|
|
5Z42
 
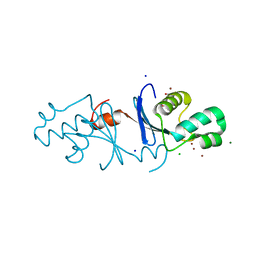 | | Aquifex aeolicus MutL endonuclease domain with three zinc ions. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DNA mismatch repair protein MutL, ... | | Authors: | Fukui, K, Yano, T. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Multiple zinc ions maintain the open conformation of the catalytic site in the DNA mismatch repair endonuclease MutL from Aquifex aeolicus
FEBS Lett., 592, 2018
|
|
1GVD
 
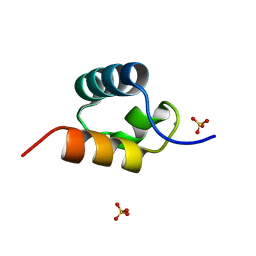 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GUU
 
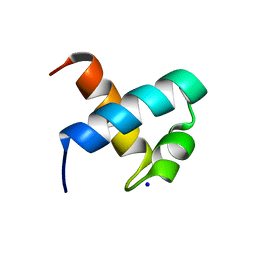 | | CRYSTAL STRUCTURE OF C-MYB R1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-01-30 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1ISR
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with Glutamate and Gadolinium Ion | | Descriptor: | GADOLINIUM ATOM, GLUTAMIC ACID, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ISS
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with an antagonist | | Descriptor: | (S)-(ALPHA)-METHYL-4-CARBOXYPHENYLGLYCINE, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1UCU
 
 | |
3A5X
 
 | |
