5ZPL
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 7 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZOZ
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 8 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPC
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 283 K (4) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPQ
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 9 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP2
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 9 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPD
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPR
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 9 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP7
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (3) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPM
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 7 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP1
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 9 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPE
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZOU
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH6 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP8
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 277 K (4) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPN
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 8 at 288 K (1) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, PHENYLACETALDEHYDE, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP0
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 8 at 288 K (2) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPF
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 6 at 288 K (3) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPT
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine at pH 10 at 288 K (2) | | Descriptor: | COPPER (II) ION, PHENYLACETALDEHYDE, Phenylethylamine oxidase, ... | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZP4
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 10 at 288 K (2) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZPH
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH6 at 293K (2) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3AGG
 
 | | X-ray analysis of lysozyme in the absence of Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGH
 
 | | X-ray analysis of lysozyme in the presence of 200 mM Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
3AGI
 
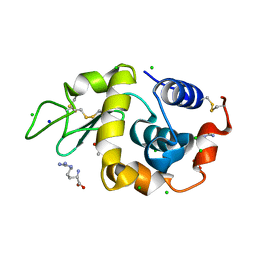 | | High resolution X-ray analysis of Arg-lysozyme complex in the presence of 500 mM Arg | | Descriptor: | ACETATE ION, ARGININE, CHLORIDE ION, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
1IOO
 
 | | CRYSTAL STRUCTURE OF NICOTIANA ALATA GEMETOPHYTIC SELF-INCOMPATIBILITY ASSOCIATED SF11-RNASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-xylopyranose-(1-2)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SF11-RNASE, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ida, K, Sato, M, Sakiyama, F, Norioka, S, Yamamoto, M, Kumasaka, T, Yamashita, E. | | Deposit date: | 2001-03-26 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The 1.55 A resolution structure of Nicotiana alata S(F11)-RNase associated with gametophytic self-incompatibility.
J.Mol.Biol., 314, 2001
|
|
1IO1
 
 | | CRYSTAL STRUCTURE OF F41 FRAGMENT OF FLAGELLIN | | Descriptor: | PHASE 1 FLAGELLIN | | Authors: | Samatey, F.A, Imada, K, Nagashima, S, Vondervisz, F, Kumasaka, T, Yamamoto, M, Namba, K. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling
Nature, 410, 2001
|
|
5X9S
 
 | | Crystal structure of fully modified H-Ras-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Ke, H, Murashima, Y, Taniguchi-Tamura, H, Miyamoto, R, Yoshikawa, Y, Kumasaka, T, Mizohata, E, Edamatsu, H, Kataoka, T. | | Deposit date: | 2017-03-09 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for intramolecular interaction of post-translationally modified H-RasGTP prepared by protein ligation
FEBS Lett., 591, 2017
|
|
