8J9D
 
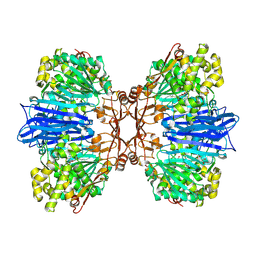 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 2024
|
|
8J9C
 
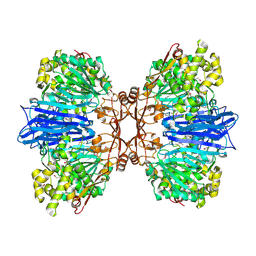 | | Crystal structure of M61 peptidase (apo-form) from Xanthomonas campestris | | Descriptor: | GLYCEROL, Putative glycyl aminopeptidase, SODIUM ION, ... | | Authors: | Yadav, P, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 2024
|
|
