6YGT
 
 | |
2FP2
 
 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
2FP1
 
 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | Chorismate mutase, LEAD (II) ION | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
2IHO
 
 | | Crystal structure of MOA, a lectin from the mushroom Marasmius oreades in complex with the trisaccharide Gal(1,3)Gal(1,4)GlcNAc | | Descriptor: | Lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Grahn, E, Askarieh, G, Holmner, A, Tateno, H, Winter, H.C, Goldstein, I.J, Krengel, U. | | Deposit date: | 2006-09-27 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of the marasmius oreades mushroom lectin in complex with a xenotransplantation epitope.
J.Mol.Biol., 369, 2007
|
|
7OKR
 
 | | Catalytic domain from the Aliivibrio salmonicida lytic polysaccharide monooxygenase AsLPMO10B | | Descriptor: | COPPER (II) ION, Chitinase B, DI(HYDROXYETHYL)ETHER | | Authors: | Cordara, G, Leitl, K.D, Loose, J.S.M, Vaaje-Kolstad, G, Krengel, U. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Chitinolytic enzymes contribute to the pathogenicity of Aliivibrio salmonicida LFI1238 in the invasive phase of cold-water vibriosis.
Bmc Microbiol., 22, 2022
|
|
7PB6
 
 | |
7PB7
 
 | |
3EF2
 
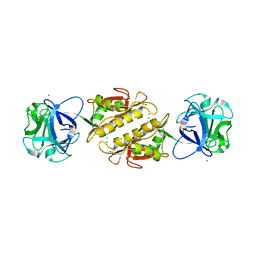 | | Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. | | Descriptor: | ACETATE ION, Agglutinin, CALCIUM ION, ... | | Authors: | Grahn, E.M, Goldstein, I.J, Krengel, U. | | Deposit date: | 2008-09-08 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of a Lectin from the Mushroom Marasmius oreades in Complex with the Blood Group B Trisaccharide and Calcium.
J.Mol.Biol., 390, 2009
|
|
3GKW
 
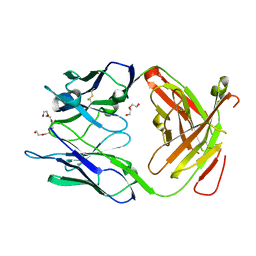 | | Crystal structure of the Fab fragment of Nimotuzumab. An anti-epidermal growth factor receptor antibody | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of the antibody Nimotuzumab, Light chain of the antibody Nimotuzumab, ... | | Authors: | Talavera, A, Friemann, R, Martinez-Fleites, C, Moreno, E, Krengel, U. | | Deposit date: | 2009-03-11 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nimotuzumab, an antitumor antibody that targets the epidermal growth factor receptor, blocks ligand binding while permitting the active receptor conformation
Cancer Res., 69, 2009
|
|
3IU4
 
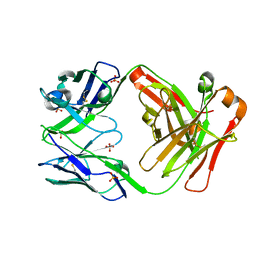 | | anti NeuGcGM3 ganglioside chimeric antibody chP3 | | Descriptor: | SULFATE ION, chP3 Fab heavy chain, chP3 Fab light chain | | Authors: | Eriksson, A, Okvist, M, Talavera, A, Krengel, U. | | Deposit date: | 2009-08-29 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of an anti-ganglioside antibody, and modelling of the functional mimicry of its NeuGc-GM3 antigen by an anti-idiotypic antibody.
Mol.Immunol., 46, 2009
|
|
5HUC
 
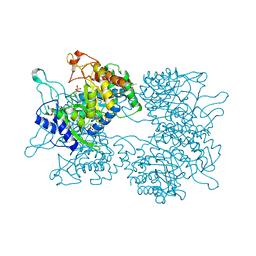 | | DAHP synthase from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, MANGANESE (II) ION, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUD
 
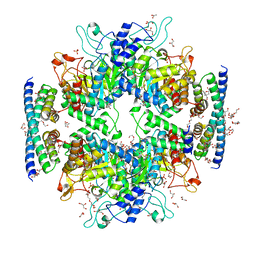 | | Non-covalent complex of and DAHP synthase and chorismate mutase from Corynebacterium glutamicum with bound transition state analog | | Descriptor: | 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUB
 
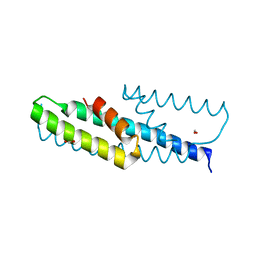 | | High-resolution structure of chorismate mutase from Corynebacterium glutamicum | | Descriptor: | Chorismate mutase, FORMIC ACID | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUE
 
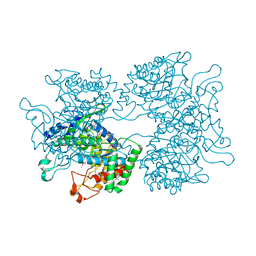 | | DAHP synthase from Corynebacterium glutamicum in complex with tryptophan | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
4NDU
 
 | |
4NDS
 
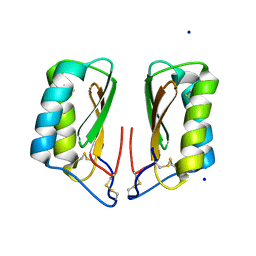 | |
4NDT
 
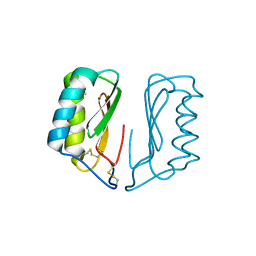 | |
4NDV
 
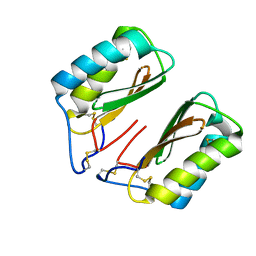 | |
1X13
 
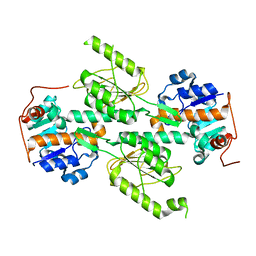 | | Crystal structure of E. coli transhydrogenase domain I | | Descriptor: | NAD(P) transhydrogenase subunit alpha | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X14
 
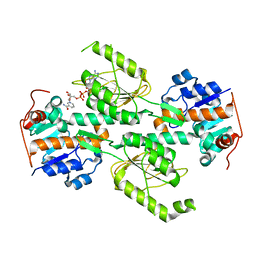 | | Crystal structure of E. coli transhydrogenase domain I with bound NAD | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X15
 
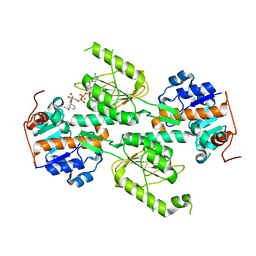 | | Crystal structure of E. coli transhydrogenase domain I with bound NADH | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
6IAL
 
 | | Porcine E.coli heat-labile enterotoxin B-pentamer in complex with Lacto-N-neohexaose | | Descriptor: | CALCIUM ION, GLYCEROL, Heat-labile enterotoxin B chain, ... | | Authors: | Heim, J.B, Heggelund, J.E, Krengel, U. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Specificity ofEscherichia coliHeat-Labile Enterotoxin Investigated by Single-Site Mutagenesis and Crystallography.
Int J Mol Sci, 20, 2019
|
|
6FFJ
 
 | |
6S2I
 
 | |
6TSN
 
 | | Marasmius oreades agglutinin (MOA), papain back.swap W208Q-Q276W variant | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
