6S53
 
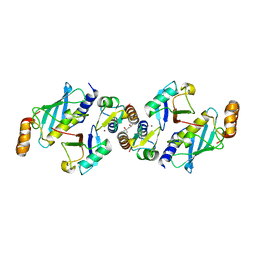 | | Crystal structure of TRIM21 RING domain in complex with an isopeptide-linked Ube2N~ubiquitin conjugate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, ... | | Authors: | Kiss, L, Boland, A, Neuhaus, D, James, L.C. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Nat Commun, 10, 2019
|
|
7BBD
 
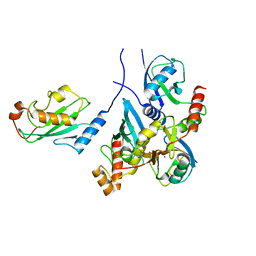 | | Crystal structure of monoubiquitinated TRIM21 RING (Ub-RING) In complex with ubiquitin charged Ube2N (Ube2N~Ub) and Ube2V2 | | Descriptor: | Polyubiquitin-B,E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Kiss, L, Neuhaus, D, James, L.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination.
Nat Commun, 12, 2021
|
|
7BBF
 
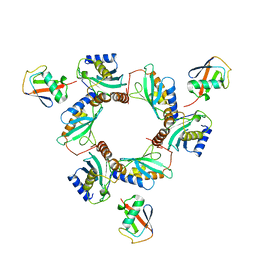 | |
8A58
 
 | |
7Q73
 
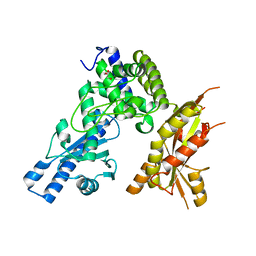 | | Structure of Pla1 apo | | Descriptor: | GLYCEROL, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q74
 
 | |
7Q72
 
 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
