4QD4
 
 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XUY
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV3
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, DI(HYDROXYETHYL)ETHER | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV6
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
5XV4
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
6M37
 
 | |
6M36
 
 | |
7WG4
 
 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG2
 
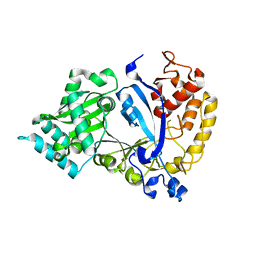 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-28 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WG1
 
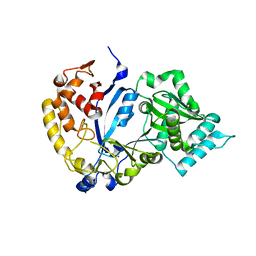 | | DVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WFX
 
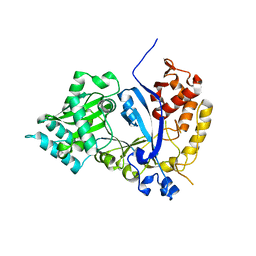 | | EVAA-KlAte1 | | Descriptor: | Arginyltransferase, ZINC ION | | Authors: | Kim, M.K, Kim, B.H, Oh, S.-J, Song, H.K. | | Deposit date: | 2021-12-27 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XZ3
 
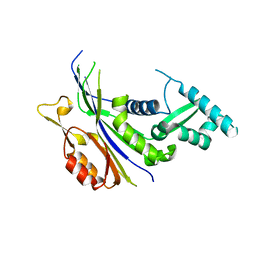 | | Crystal structure of the Type I-B CRISPR-associated protein, Csh2 from Thermobaculum terrenum | | Descriptor: | CRISPR-associated protein, Csh2 family | | Authors: | Seo, P.W, Gu, D.H, Park, S.Y, Kim, J.S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural characterization of the type I-B CRISPR Cas7 from Thermobaculum terrenum.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
6TY0
 
 | |
6TY2
 
 | |
6VER
 
 | | Human insulin analog: [GluB10,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
6VBP
 
 | |
6VBO
 
 | |
6VBQ
 
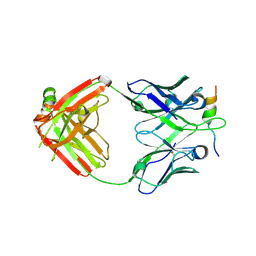 | |
6VES
 
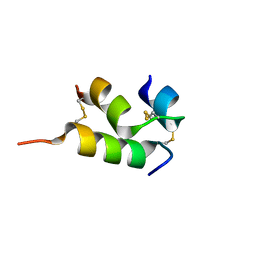 | | Human insulin analog: [GluB10,HisA8,ArgA9]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mini-Ins: A Minimal, Bioactive Insulin Analog with Alternative Binding Modes
To Be Published
|
|
6VQN
 
 | | Co-crystal structure of human PD-L1 complexed with Compound A | | Descriptor: | N,N'-(2,2'-dimethyl[1,1'-biphenyl]-3,3'-diyl)bis(5-{[(2-hydroxyethyl)amino]methyl}pyridine-2-carboxamide), Programmed cell death 1 ligand 1 | | Authors: | White, A, Lakshminarasimhan, D, Leo, C, Suto, R.K. | | Deposit date: | 2020-02-05 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1.
Nat Commun, 12, 2021
|
|
5Y16
 
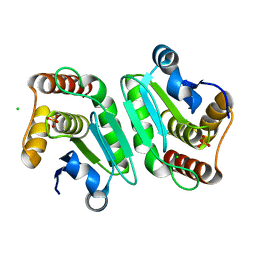 | | Crystal structure of human DUSP28(Y102H) | | Descriptor: | CHLORIDE ION, Dual specificity phosphatase 28, PHOSPHATE ION | | Authors: | Ku, B, Kim, M, Kim, S.J, Ryu, S.E. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural and biochemical analysis of atypically low dephosphorylating activity of human dual-specificity phosphatase 28
PLoS ONE, 12, 2017
|
|
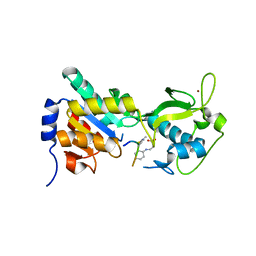
3RIG
 
 | |
3RIY
 
 | |
5XAC
 
 | | CLIR - LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XAE
 
 | | mutNLIR_LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
