6ZWV
 
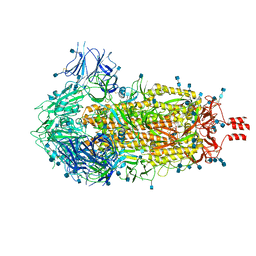 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
8XAB
 
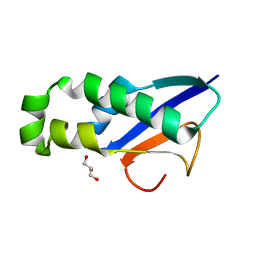 | |
6XL0
 
 | | Caulobacter crescentus FljK filament | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
6XKY
 
 | | Caulobacter crescentus FljK filament, straightened | | Descriptor: | Flagellin | | Authors: | Montemayor, E.J, Ploscariu, N.T, Sanchez, J.C, Parrell, D, Dillard, R.S, Shebelut, C.W, Ke, Z, Guerrero-Ferreira, R.C, Wright, E.R. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flagellar Structures from the Bacterium Caulobacter crescentus and Implications for Phage phi CbK Predation of Multiflagellin Bacteria
J.Bacteriol., 203, 2021
|
|
3RHY
 
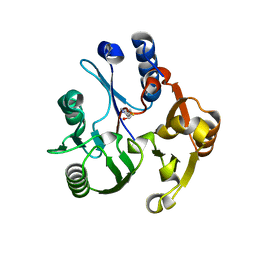 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
7EQ4
 
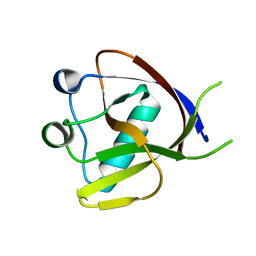 | | Crystal Structure of the N-terminus of Nonstructural protein 1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Liu, Y, Ke, Z, Hu, H, Zhao, K, Xiao, J, Xia, Y, Li, Y. | | Deposit date: | 2021-04-29 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Microbiol Spectr, 9, 2021
|
|
7OVR
 
 | | Mature HIV-1 matrix structure | | Descriptor: | HIV-1 matrix, MYRISTIC ACID, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
7OVQ
 
 | | Immature HIV-1 matrix structure | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
8BQE
 
 | | In situ structure of the Caulobacter crescentus S-layer | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose, CALCIUM ION, S-layer protein rsaA | | Authors: | von Kuegelgen, A, Bharat, T. | | Deposit date: | 2022-11-21 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
3VB3
 
 | | Crystal structure of SARS-CoV 3C-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, DI(HYDROXYETHYL)ETHER | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB5
 
 | | Crystal structure of SARS-CoV 3C-like protease with C4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C4Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB4
 
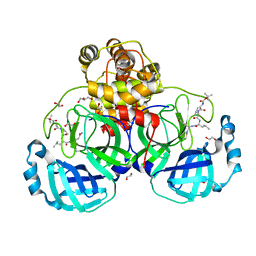 | | Crystal structure of SARS-CoV 3C-like protease with B4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, B4Z inhibitor, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB7
 
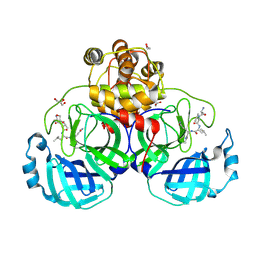 | | Crystal structure of SARS-CoV 3C-like protease with M4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, GLYCEROL, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
3VB6
 
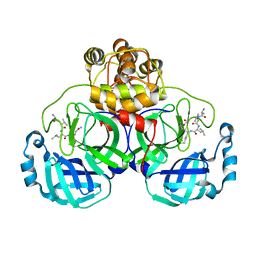 | | Crystal structure of SARS-CoV 3C-like protease with C6Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, C6Z inhibitor | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
7V3T
 
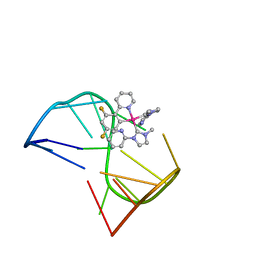 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
8BSH
 
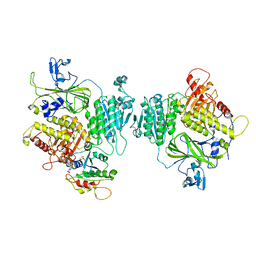 | | COPII inner coat | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein transport protein SEC23, ... | | Authors: | Zanetti, G, Pyle, E.W. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.
Elife, 11, 2022
|
|
6ZOZ
 
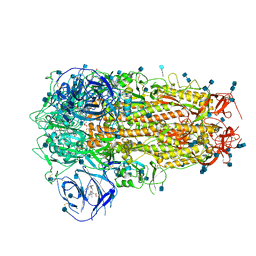 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOX
 
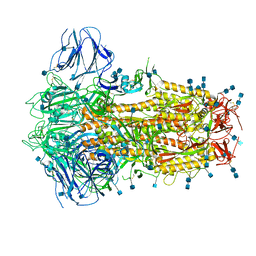 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP2
 
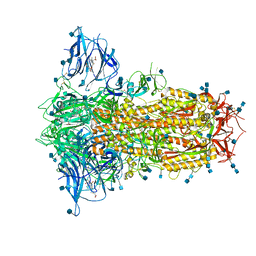 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOY
 
 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP0
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP1
 
 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5YJO
 
 | |
6IJL
 
 | |
8JVD
 
 | |
