4XCM
 
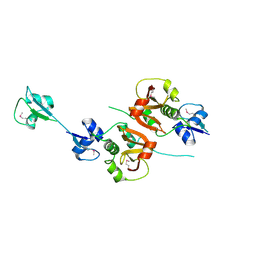 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2L60
 
 | |
4UZ3
 
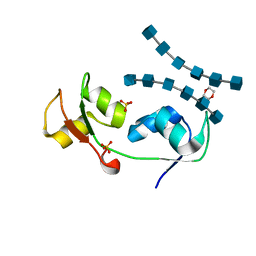 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X1N
 
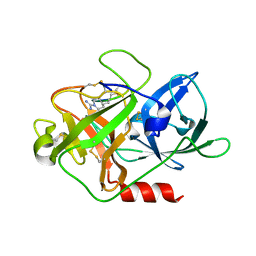 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X1Q
 
 | | The crystal structure of mupain-1 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X0W
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1S
 
 | | The crystal structure of mupain-1-16-D9A in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X1R
 
 | | The crystal structure of mupain-1-12 in complex with murinised human uPA at pH7.4 | | Descriptor: | 1-phenylguanidine, Urokinase-type plasminogen activator, mupain-1-12 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
6XWE
 
 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
3OY6
 
 | |
3OY5
 
 | |
3OX7
 
 | | The crystal structure of uPA complex with peptide inhibitor MH027 at pH4.6 | | Descriptor: | MH027, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Andreasen, P.A, Huang, M.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The binding mechanism of a peptidic cyclic serine protease inhibitor
J.Mol.Biol., 412, 2011
|
|
4ZHM
 
 | |
4ZHL
 
 | |
4UZ2
 
 | |
7AU7
 
 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BAX
 
 | | Crystal structure of LYS11 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | Authors: | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-12-16 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4EFX
 
 | |
3U4N
 
 | | A novel covalently linked insulin dimer | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Norrman, M, Vinther, T.N. | | Deposit date: | 2011-10-10 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel covalently linked insulin dimer engineered to investigate the function of insulin dimerization.
Plos One, 7, 2012
|
|
