8P60
 
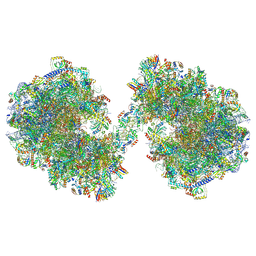 | | Spraguea lophii ribosome dimer | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2023-05-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (14.3 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
7ZCX
 
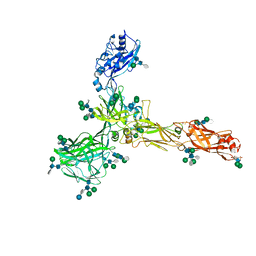 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-03-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
7ZUG
 
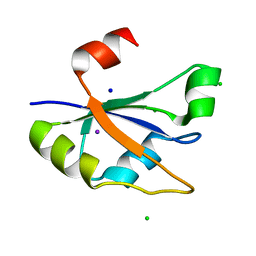 | | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain | | Descriptor: | CHLORIDE ION, Heterogeneous nuclear ribonucleoprotein H, N-terminally processed, ... | | Authors: | Winter, N, Kumar, M, Isupov, M.N, Wiener, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.075 Å) | | Cite: | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain
To Be Published
|
|
7NW1
 
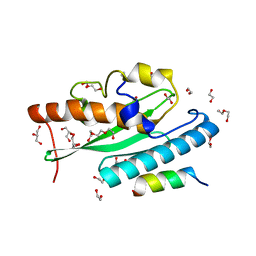 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVJ
 
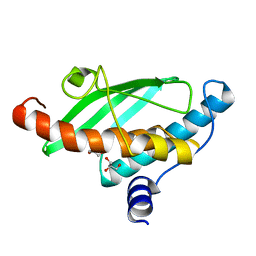 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
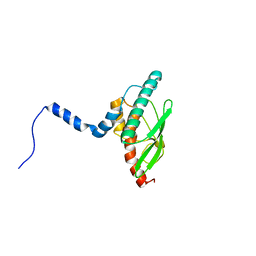 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NZQ
 
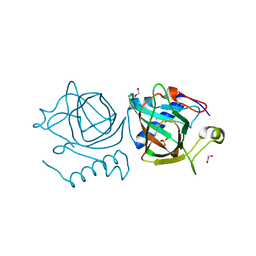 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
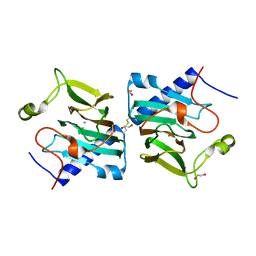 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
5AOB
 
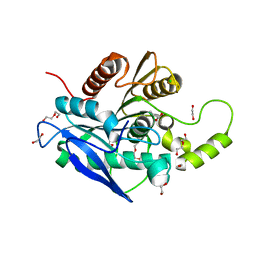 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AIG
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample- Valpromide complex | | Descriptor: | 2-PROPYLPENTANAMIDE, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AII
 
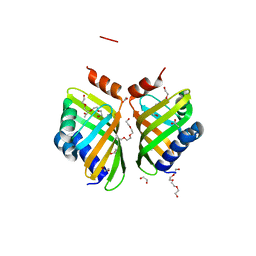 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-PEG complex | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AIH
 
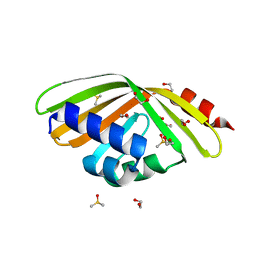 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. CH55-sample-Native | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AOC
 
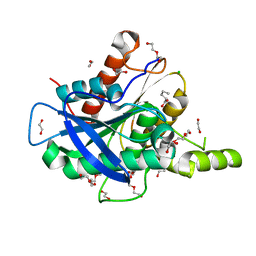 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AOA
 
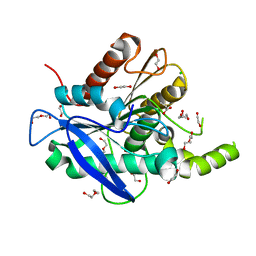 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AIF
 
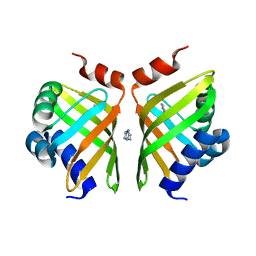 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample-Native | | Descriptor: | IMIDAZOLE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5AO9
 
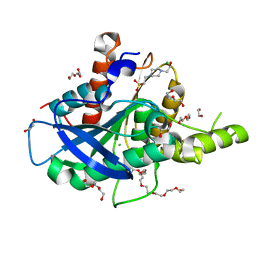 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
1T9H
 
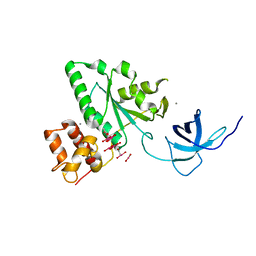 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
7P7W
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PA1
 
 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PVI
 
 | | dTDP-sugar epimerase | | Descriptor: | CITRATE ANION, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
