4QTS
 
 | |
4DWX
 
 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Basic endochitinase C, SULFATE ION, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-27 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
4DYG
 
 | | Crystal Structure of a Family GH-19 Chitinase from rye seeds in complex with (GlcNAc)4 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic endochitinase C, ... | | Authors: | Numata, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2012-02-29 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and chitin oligosaccharide-binding mode of a 'loopful' family GH19 chitinase from rye, Secale cereale, seeds
Febs J., 279, 2012
|
|
6AE2
 
 | |
6AE1
 
 | |
3W2W
 
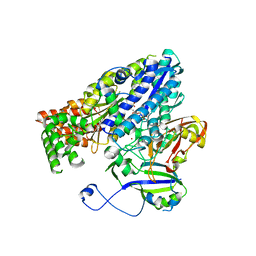 | | Crystal structure of the Cmr2dHD-Cmr3 subcomplex bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2012-12-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Cmr2-Cmr3 Subcomplex in the CRISPR-Cas RNA Silencing Effector Complex.
J.Mol.Biol., 425, 2013
|
|
3W2V
 
 | |
3X1L
 
 | |
2ZXI
 
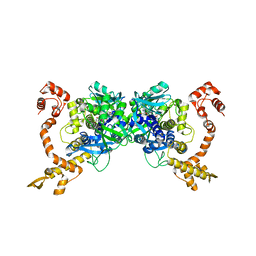 | | Structure of Aquifex aeolicus GidA in the form II crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
2ZXH
 
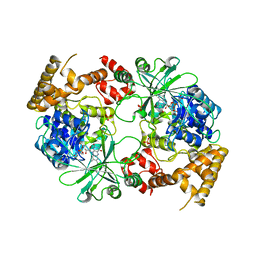 | | Structure of Aquifex aeolicus GidA in the form I crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
3AMU
 
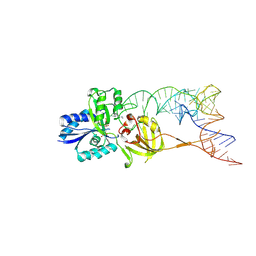 | |
3AMT
 
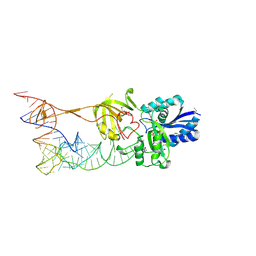 | | Crystal structure of the TiaS-tRNA(Ile2)-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, RNA (78-MER) | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2010-08-23 | | Release date: | 2011-10-12 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of tRNA agmatinylation essential for AUA codon decoding
Nat.Struct.Mol.Biol., 18, 2011
|
|
3AU7
 
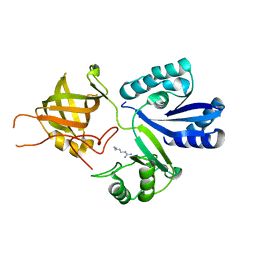 | |
