5J3G
 
 | |
3U3I
 
 | | A RNA binding protein from Crimean-Congo hemorrhagic fever virus | | Descriptor: | Nucleocapsid protein | | Authors: | Guo, Y, Wang, W.M, Ji, W, Deng, M, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-10-06 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D36
 
 | |
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
2P9C
 
 | |
2PA3
 
 | |
2P9G
 
 | |
2P9E
 
 | |
4LPG
 
 | | Crystal structure of human FPPS in complex with CL01131 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methanediyl)bis(phosphonic acid), Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
4LPH
 
 | | Crystal structure of human FPPS in complex with CL03093 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl)phosphonic acid, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
3OEV
 
 | | Structure of yeast 20S open-gate proteasome with Compound 25 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(benzyloxy)-N-[(2S,3R)-3-hydroxy-1-{[(2S)-1-{[(3-methylthiophen-2-yl)methyl]amino}-1-oxo-4-phenylbutan-2-yl]amino}-1-oxobutan-2-yl]benzamide, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OEU
 
 | | Structure of yeast 20S open-gate proteasome with Compound 24 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-{(2S)-1-[(2-chlorobenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[3-(2-methylphenyl)propanoyl]-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8HM0
 
 | | F8-A22-E4 complex of MPXV in trimeric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
8HLZ
 
 | | F8-A22-E4 complex of MPXV in hexameric form | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Li, Y.N, Shen, Y.P, Hu, Z.W, Yan, R.H. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the assembly of the DNA polymerase holoenzyme from a monkeypox virus variant.
Sci Adv, 9, 2023
|
|
5W3X
 
 | | Crystal structure of PopP2 in complex with IP6, AcCoA and the WRKY domain of RRS1-R . | | Descriptor: | ACETYL COENZYME *A, Disease resistance protein RRS1, GLYCEROL, ... | | Authors: | Zhang, Z.M, Gao, L, Song, J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
5W3T
 
 | | Crystal structure of PopP2 in complex with IP6 | | Descriptor: | GLYCEROL, INOSITOL HEXAKISPHOSPHATE, PopP2 protein | | Authors: | Song, J, Zhang, Z.M. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
5W40
 
 | |
5W3Y
 
 | |
5WYZ
 
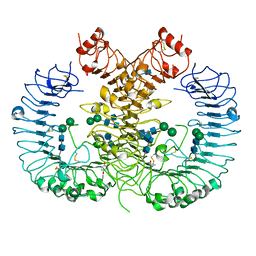 | | Crystal structure of human TLR8 in complex with CU-CPT9b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-methyl-4-oxidanyl-phenyl)quinolin-7-ol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition of TLR8 through stabilization of its resting state
Nat. Chem. Biol., 14, 2018
|
|
8JLX
 
 | | CCHFV envelope protein Gc in complex with Gc13 | | Descriptor: | Glycoprotein C,CCHFV Gc fusion loops, Mouse antibody Gc13 heavy chain, Mouse antibody Gc13 light chain | | Authors: | Chong, T, Cao, S. | | Deposit date: | 2023-06-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.
Plos Pathog., 20, 2024
|
|
8JKD
 
 | |
