4XXI
 
 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XXK
 
 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | Descriptor: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | Authors: | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | Deposit date: | 2015-01-30 | | Release date: | 2015-12-16 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6Z68
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2021-06-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6Z69
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
4Q5O
 
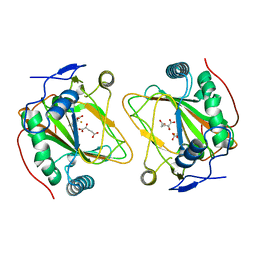 | | Crystal structure of EctD from S. alaskensis with 2-oxoglutarate and 5-hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, Ectoine hydroxylase, ... | | Authors: | Hoeppner, A, Widderich, N, Bremer, E, Smits, S.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
3JYP
 
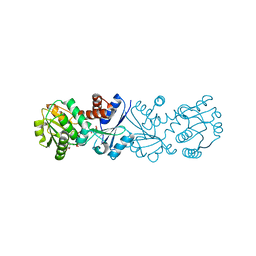 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYO
 
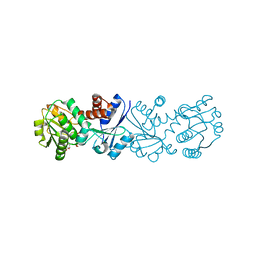 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Niefind, K, Schomburg, D. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYQ
 
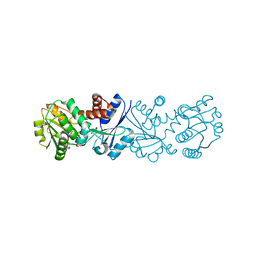 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with shikimate and NADH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
4MHU
 
 | | Crystal structure of EctD from S. alaskensis with bound Fe | | Descriptor: | Ectoine hydroxylase, FE (III) ION, N-DODECYL-N,N-DIMETHYLGLYCINATE | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4MHR
 
 | | Crystal structure of EctD from S. alaskensis in its apoform | | Descriptor: | Ectoine hydroxylase | | Authors: | Widderich, N, Hoeppner, A, Pittelkow, M, Heider, J, Smits, S.H, Bremer, E. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
8B6E
 
 | | crystal structure of the DNA-binding short chromatophore-targeted protein sCTP-23166 from Paulinella chromatophora | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, sCTP-23166 | | Authors: | Macorano, L, Applegate, V, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | DNA-binding and protein structure of nuclear factors likely acting in genetic information processing in the Paulinella chromatophore.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6C7N
 
 | | Monoclinic form of malic enzyme from sorghum at 2 angstroms resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Malic enzyme, ... | | Authors: | Trajtenberg, F, Alvarez, C, Buschiazzo, A. | | Deposit date: | 2018-01-23 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular adaptations of NADP-malic enzyme for its function in C4photosynthesis in grasses.
Nat.Plants, 5, 2019
|
|
7BBR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T | | Descriptor: | 2-KETO-3-DEOXYGLUCONATE, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCP
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with gluconate | | Descriptor: | D-gluconic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCO
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with D-foconate | | Descriptor: | (2R,3S,4S,5S)-2,3,4,5-tetrahydroxyhexanoic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with galactonate | | Descriptor: | L-galactonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
7BCN
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with Xylonic acid | | Descriptor: | D-xylonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
5LU4
 
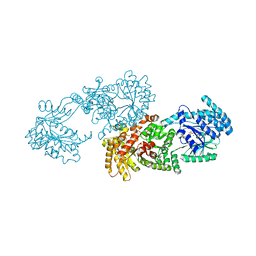 | | C4-type pyruvate phosphate dikinase: conformational intermediate of central domain in the swiveling mechanism | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Trapped intermediate state of plant pyruvate phosphate dikinase indicates substeps in catalytic swiveling domain mechanism.
Protein Sci., 26, 2017
|
|
6HRK
 
 | | Structure of a far-red fluorescent biliprotein derived from a far-red induced allophycocyanin F subunit from a thermophilic cyanobacterium Chroococcidiopsis thermalis | | Descriptor: | Allophycocyanin beta-18 subunit apoprotein, BILIVERDINE IX ALPHA | | Authors: | Hou, Y.-N, Hoeppner, A, Ding, W.-L, Gaertner, W, Zhao, K.-H. | | Deposit date: | 2018-09-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Control of a far-red/near-infrared spectral switch in an artificial fluorescent biliprotein derived from allophycocyanin
Protein Sci., Suppl.: Diskette Appendix To V. , No. , [Month], Filename:, 31, 2022
|
|
8OR7
 
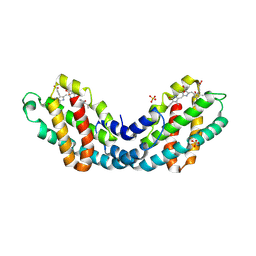 | | Structure of a far-red induced allophycocyanin from Chroococcidiopsis thermalis sp. PCC 7203 | | Descriptor: | Allophycocyanin beta subunit apoprotein, PHYCOCYANOBILIN, POTASSIUM ION, ... | | Authors: | Zhou, L.J, Hoeppner, A, Wang, Y.Q, Zhao, K.H. | | Deposit date: | 2023-04-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and biochemical analyses of a far-red allophycocyanin to address the mechanism of the super-red-shift.
Photosynth.Res., 2024
|
|
7PZE
 
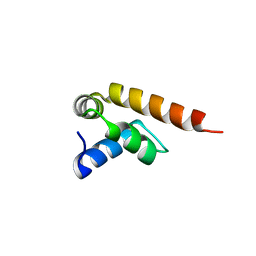 | | MademoiseLLE domain 2 of Rrm4 from Ustilago maydis | | Descriptor: | Chromosome 8, whole genome shotgun sequence | | Authors: | Devans, S, Schott-Verdugo, s, Muentjes, K, Olgeiser, L, Reiners, J, Schmitt, L, Hoeppner, A, Smits, S.H, Gohlke, H, Feldbruegge, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A MademoiseLLE domain binding platform links the key RNA transporter to endosomes.
Plos Genet., 18, 2022
|
|
6SBN
 
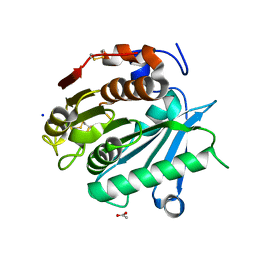 | | Polyester hydrolase PE-H of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, SODIUM ION, polyester hydrolase | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SCD
 
 | | Polyester hydrolase PE-H Y250S mutant of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
5BXX
 
 | | Crystal structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-09 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
5BY5
 
 | | High resolution structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase, S-1,2-PROPANEDIOL | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-10 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
