6BD4
 
 | | Crystal structure of human apo-Frizzled4 receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Frizzled-4/Rubredoxin chimeric protein, OLEIC ACID, ... | | Authors: | Yang, S, Wu, Y, Pu, M, Chen, Y, Dong, S, Guo, Y, Han, G.Y, Stevens, R.C, Zhao, S, Xu, F. | | Deposit date: | 2017-10-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Frizzled 4 receptor in a ligand-free state.
Nature, 560, 2018
|
|
5JHP
 
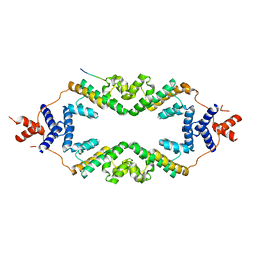 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JGC
 
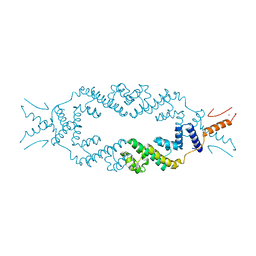 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A, L130A, L179A and I195A mutant | | Descriptor: | Protein TPR1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-20 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JA5
 
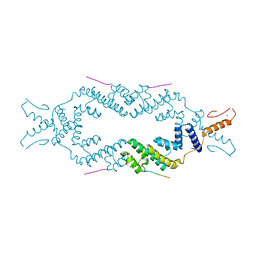 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5J9K
 
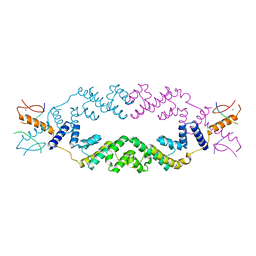 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, ZINC ION, rice D53 peptide 794-808 | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
4RDD
 
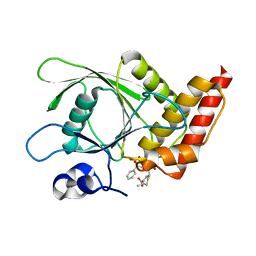 | | Co-crystal structure of SHP2 in complex with a Cefsulodin derivative | | Descriptor: | 1-({(2R)-4-carboxy-2-[(R)-carboxy{[(2R)-2-phenyl-2-sulfoacetyl]amino}methyl]-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.Y, Yu, Z.H, He, R, Zhang, R.Y. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-01 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Exploring the Existing Drug Space for Novel pTyr Mimetic and SHP2 Inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
8ENX
 
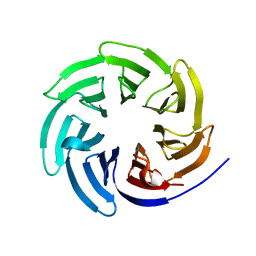 | |
8ENY
 
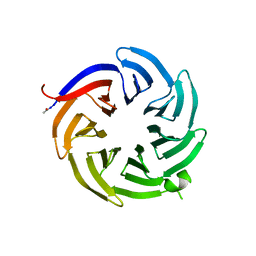 | |
8ENZ
 
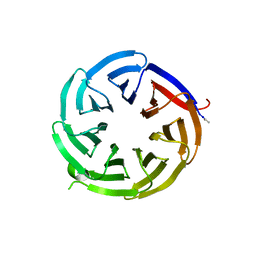 | |
8EO0
 
 | |
8ENS
 
 | |
8ENW
 
 | |
4N9L
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9K
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N92
 
 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
5H4B
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
7LKI
 
 | |
5GHY
 
 | |
5GHZ
 
 | |
5GHX
 
 | | Crystal structure of beta-lactamase PenP mutant-E166H | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2016-06-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Snapshots of Class A beta-Lactamase Catalysis Reveal Structural Changes That Facilitate beta-Lactam Hydrolysis
J. Biol. Chem., 292, 2017
|
|
4HVI
 
 | |
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
5TTO
 
 | | X-ray crystal structure of PPARgamma in complex with SR1643 | | Descriptor: | 4-bromo-N-{3,5-dichloro-4-[(quinolin-3-yl)oxy]phenyl}-2,5-difluorobenzene-1-sulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P, Kamenecka, T, Abell, A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPAR gamma-Targeted Antidiabetics.
J. Med. Chem., 60, 2017
|
|
4HVG
 
 | |
4HVH
 
 | |
