6OGL
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGV
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in apo state | | Descriptor: | Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6OGS
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OYD
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-004 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGR
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
6OYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-002 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
1A3G
 
 | | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Sato, M, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1998-01-21 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Escherichia coli branched-chain amino acid aminotransferase at 2.5 A resolution.
J.Biochem.(Tokyo), 121, 1997
|
|
1AMR
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
1AMS
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLUTARIC ACID | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
1AMQ
 
 | | X-RAY CRYSTALLOGRAPHIC STUDY OF PYRIDOXAMINE 5'-PHOSPHATE-TYPE ASPARTATE AMINOTRANSFERASES FROM ESCHERICHIA COLI IN THREE FORMS | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Miyahara, I, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 1994-07-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic study of pyridoxamine 5'-phosphate-type aspartate aminotransferases from Escherichia coli in three forms.
J.Biochem.(Tokyo), 116, 1994
|
|
4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2N
 
 | | Crystal structure of MHPCO, Y270F mutant | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2Q
 
 | | structure of MHPCO-5HN complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridiine-5-carboxylic acid oxygenase
To be Published
|
|
4JY3
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-pyridoxic acid bound form | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-4-(hydroxymethyl)-6-methylpyridine-3-carboxylic acid, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-29 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2P
 
 | | Tetrameric form of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4H2R
 
 | | Structure of MHPCO Y270F mutant, 5-hydroxynicotinic acid complex | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxypyridine-3-carboxylic acid, BETA-MERCAPTOETHANOL, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Yagi, T. | | Deposit date: | 2012-09-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4JY2
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, native and unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Kamitori, S, Hayashi, H, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
6D0E
 
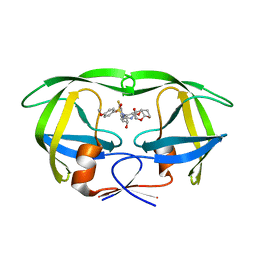 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-084-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
6D0D
 
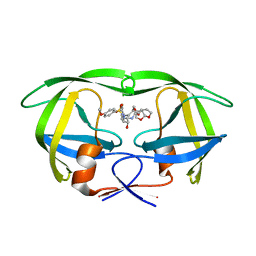 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-087-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4V
 
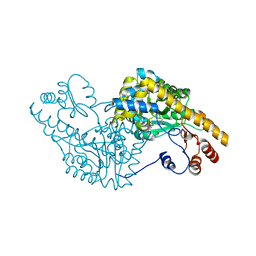 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G7W
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
5COK
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5ZOX
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by ethylamine at pH 7 at 288 K (1) | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Baba, S, Kawano, Y, Hayashi, H, Yano, T, Tanizawa, K, Kumasaka, T, Yamamoto, M, Okajima, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | In crystallothermodynamic analysis of conformational change of the topaquinone cofactor in bacterial copper amine oxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
