6JG9
 
 | |
6JG8
 
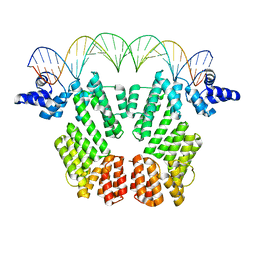 | | Crystal structure of AimR in complex with DNA | | Descriptor: | AimR transcriptional regulator, DNA (31-MER) | | Authors: | Guan, Z.Y, Pei, K, Zou, T.T. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
6JG5
 
 | | Crystal structure of AimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Guan, Z.Y, Pei, K, Zou, T.T. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
7V9X
 
 | |
7YTJ
 
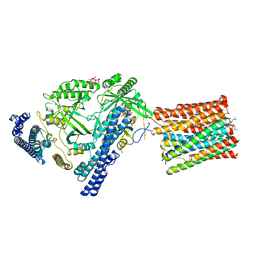 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
8J5D
 
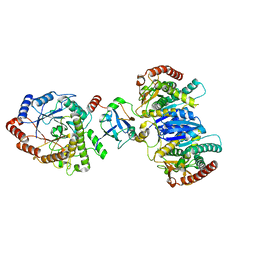 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
7D5K
 
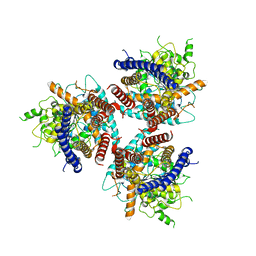 | | CryoEM structure of cotton cellulose synthase isoform 7 | | Descriptor: | Cellulose synthase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guan, Z.Y, Xue, Y, Yin, P, Zhang, X.L. | | Deposit date: | 2020-09-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum.
Plant Biotechnol J, 19, 2021
|
|
7D3T
 
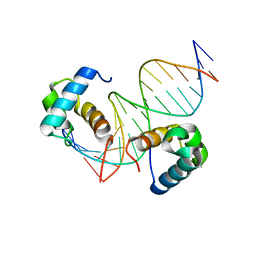 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
5JCB
 
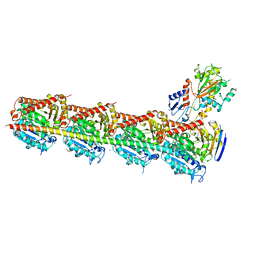 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | Descriptor: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Guan, Z, Zhao, W, Yin, P. | | Deposit date: | 2016-04-14 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
8W5K
 
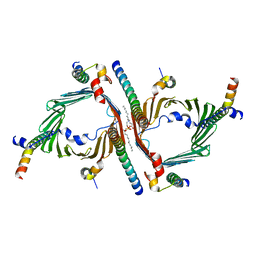 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5J
 
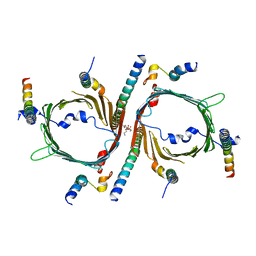 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
6LNI
 
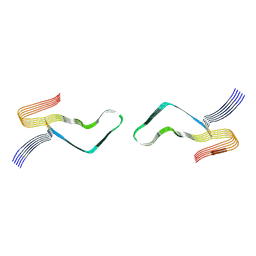 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2020-06-24 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6M79
 
 | |
6LCT
 
 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6KVO
 
 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-09-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LCM
 
 | | Crystal structure of chloroplast resolvase ZmMOC1 with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ZmMoc1 | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6L77
 
 | | Crystal structure of MS5 from Brassica napus | | Descriptor: | MS5a | | Authors: | Wang, X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the meiosis-related protein MS5 reveals non-canonical papain enhancement by cystatin-like folds.
Febs Lett., 594, 2020
|
|
8H0N
 
 | | Crystal structure of the human METTL1-WDR4 complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
8GQE
 
 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
6KVN
 
 | |
5XYB
 
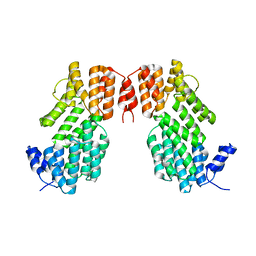 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
5YEJ
 
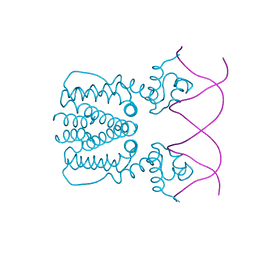 | | Crystal structure of BioQ with its naturel double-stranded DNA operator | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*AP*AP*CP*AP*CP*CP*GP*TP*TP*CP*AP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*TP*GP*AP*AP*CP*GP*GP*TP*GP*TP*TP*CP*AP*GP*GP*T)-3'), TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5YEK
 
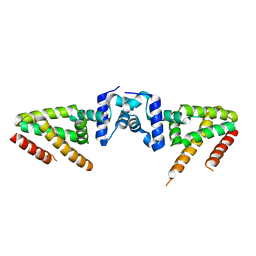 | | Crystal structure of BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5YGU
 
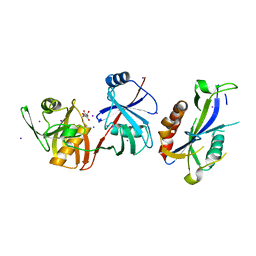 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
