1BV1
 
 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | BET V 1 | | Authors: | Gajhede, M, Osmark, P, Poulsen, F.M, Ipsen, H, Larson, J.N, Joostvan, R.J, Schou, C, Lowenstein, H, Spangfort, M.D. | | Deposit date: | 1997-07-08 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
1ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C1A | | Descriptor: | CALCIUM ION, PEROXIDASE C1A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gajhede, M, Schuller, D.J, Henriksen, A, Smith, A.T, Poulos, T.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of horseradish peroxidase C at 2.15 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
1PSP
 
 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
6ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C COMPLEX WITH FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-23 | | Release date: | 2000-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
6RUQ
 
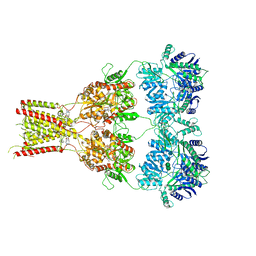 | | Structure of GluA2cryst in complex the antagonist ZK200775 and the negative allosteric modulator GYKI53655 at 4.65 A resolution | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krintel, C, Venskutonyte, R, Mirza, O.A, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.65 Å) | | Cite: | Binding of a negative allosteric modulator and competitive antagonist can occur simultaneously at the ionotropic glutamate receptor GluA2.
Febs J., 288, 2021
|
|
6F6D
 
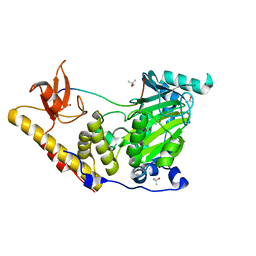 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
4UM3
 
 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3920 | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
4UM1
 
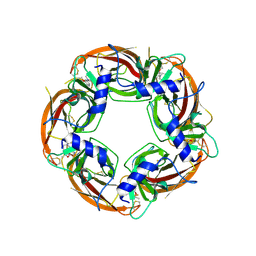 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3573 | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
5JT8
 
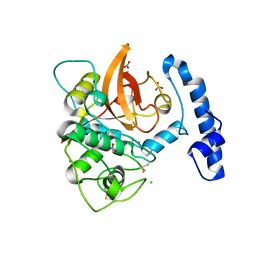 | | Structural basis for the limited antibody cross reactivity between the mite allergens Blo t 1 and Der p 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Blo t 1 allergen, ... | | Authors: | Meno, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the mite allergen Blo t 1 explains the limited antibody cross-reactivity to Der p 1.
Allergy, 72, 2017
|
|
1ZS2
 
 | | Amylosucrase Mutant E328Q in a ternary complex with sucrose and maltoheptaose | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, van der Veen, B.A, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the Glu328Gln mutant of Neisseria polysaccharea amylosucrase in complex with sucrose and maltoheptaose
BIOCATAL.BIOTRANSFOR., 24, 2006
|
|
4ATJ
 
 | | DISTAL HEME POCKET MUTANT (H42E) OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (PEROXIDASE C1A), ... | | Authors: | Meno, K, Jennings, S, Smith, A.T, Henriksen, A, Gajhede, M. | | Deposit date: | 1999-04-19 | | Release date: | 2002-10-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5CC2
 
 | | STRUCTURE OF THE LIGAND-BINDING DOMAIN OF THE IONOTROPIC GLUTAMATE RECEPTOR-LIKE GLUD2 IN COMPLEX WITH 7-CKA | | Descriptor: | 7-Chlorokynurenic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Pharmacology and Structural Analysis of Ligand Binding to the Orthosteric Site of Glutamate-Like GluD2 Receptors.
Mol.Pharmacol., 89, 2016
|
|
1MW2
 
 | | Amylosucrase soaked with 100mM sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MW3
 
 | | Amylosucrase soaked with 1M sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MVY
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MW1
 
 | | Amylosucrase soaked with 14mM sucrose. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
1MW0
 
 | | Amylosucrase mutant E328Q co-crystallized with maltoheptaose then soaked with maltoheptaose. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
3S9E
 
 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding site and interlobe interactions of the ionotropic glutamate receptor GluK3 ligand binding domain revealed by high resolution crystal structure in complex with (S)-glutamate.
J.Struct.Biol., 176, 2011
|
|
1KZM
 
 | | Distal Heme Pocket Mutant (R38S/H42E) of Recombinant Horseradish Peroxidase C (HRP C). | | Descriptor: | CACODYLATE ION, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meno, K, Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7OFE
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 2-[(9-oxidanylidenefluoren-4-yl)carbonylamino]ethanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OF9
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, [(4S)-6,7-dihydro-4H-thieno[3,2-c]pyran-4-yl]methanamine | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFB
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFF
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | (2~{R})-2-[(9-oxidanylidenefluoren-4-yl)carbonylamino]-2-phenyl-ethanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
7OFC
 
 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
