2VN5
 
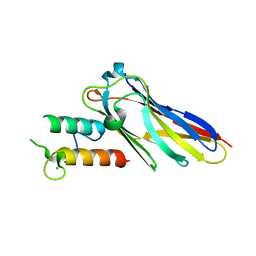 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2VN6
 
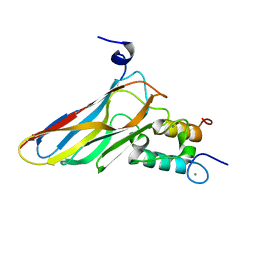 | | The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner | | Descriptor: | CALCIUM ION, ENDOGLUCANASE A, SCAFFOLDING PROTEIN | | Authors: | Pinheiro, B.A, Prates, J.A.M, Proctor, M.R, Gilbert, H.J, Davies, G.J, Money, V.A, Martinez-Fleites, C, Bayer, E.A, Fontes, C.M.G.A, Fierobe, H.P. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Clostridium Cellulolyticum Dockerin Displays a Dual Binding Mode for its Cohesin Partner.
J.Biol.Chem., 283, 2008
|
|
2WAO
 
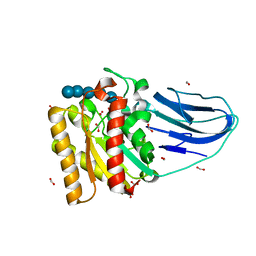 | | Structure of a family two carbohydrate esterase from Clostridium thermocellum in complex with cellohexaose | | Descriptor: | ENDOGLUCANASE E, FORMIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-10 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2W47
 
 | | Clostridium thermocellum CBM35 in complex with delta-4,5- anhydrogalacturonic acid | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, LIPOLYTIC ENZYME, ... | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W87
 
 | | Xyl-CBM35 in complex with glucuronic acid containing disaccharide. | | Descriptor: | CALCIUM ION, ESTERASE D, UNKNOWN LIGAND, ... | | Authors: | Montainer, C, Bueren, A.L.v, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WAA
 
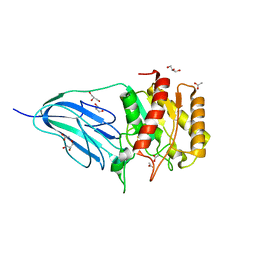 | | Structure of a family two carbohydrate esterase from Cellvibrio japonicus | | Descriptor: | ACETATE ION, GLYCEROL, XYLAN ESTERASE, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2W46
 
 | | CBM35 from Cellvibrio japonicus Abf62 | | Descriptor: | CALCIUM ION, ESTERASE D, SODIUM ION | | Authors: | Montainer, C, Lammerts van Bueren, A, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-01-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VPT
 
 | | Clostridium thermocellum family 3 carbohydrate esterase | | Descriptor: | CALCIUM ION, LIPOLYTIC ENZYME | | Authors: | Correia, M.A.S, Prates, J.A.M, Bras, J, Fontes, C.M.G.A, Newman, J.A, Lewis, R.J, Gilbert, H.J, Flint, J.E. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-06 | | Last modified: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Cellulosomal Family 3 Carbohydrate Esterase from Clostridium Thermocellum Provides Insights Into the Mechanism of Substrate Recognition
J.Mol.Biol., 379, 2008
|
|
2WAB
 
 | | Structure of an active site mutant of a family two carbohydrate esterase from Clostridium thermocellum in complex with celluohexase | | Descriptor: | ENDOGLUCANASE E, GLYCEROL, IODIDE ION, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
6SAB
 
 | | M-BUTX-Ptr1a (Parabuthus transvaalicus) | | Descriptor: | M-BUTX-Ptr1a | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
7O8C
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
6FOP
 
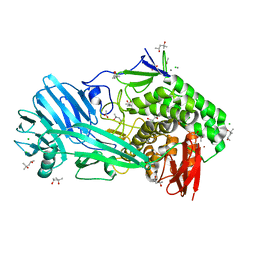 | | Glycoside hydrolase family 81 from Clostridium thermocellum (CtLam81A), Mutant E515A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Correia, M.A.S.C, Carvalho, A.L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel insights into the degradation of beta-1,3-glucans by the cellulosome of Clostridium thermocellum revealed by structure and function studies of a family 81 glycoside hydrolase.
Int.J.Biol.Macromol., 117, 2018
|
|
2M1J
 
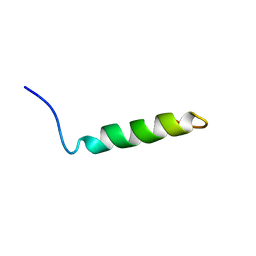 | | Ovine Doppel Signal peptide (1-30) | | Descriptor: | Prion-like protein doppel | | Authors: | Pimenta, J, Viegas, A, Sardinha, J, Santos, A, Cantante, C, Dias, F.M.V, Soares, R, Cabrita, E.J, Fontes, C.M.G.A, Prates, J.A.M, Pereira, R.M.L.N. | | Deposit date: | 2012-11-28 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and SRP54M predicted interaction of the N-terminal sequence (1-30) of the ovine Doppel protein.
Peptides, 49C, 2013
|
|
1UXZ
 
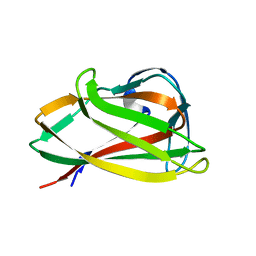 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
7NOX
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus in complex with mixed-linked gluco-nonasaccharide | | Descriptor: | AZIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
4DH2
 
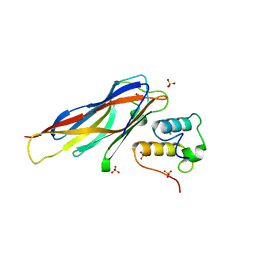 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
5T87
 
 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
1UZ4
 
 | | Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, GLYCEROL, MAN5A, ... | | Authors: | Vincent, F, Davies, G.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Common Inhibition of Both Beta-Glucosidases and Beta-Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
6R31
 
 | | Family 11 Carbohydrate-Binding Module from Clostridium thermocellum in complex with beta-1,3-1,4-mixed-linked tetrasaccharide | | Descriptor: | CALCIUM ION, Endoglucanase H, PHOSPHATE ION, ... | | Authors: | Ribeiro, D.O, Carvalho, A.L. | | Deposit date: | 2019-03-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the preferential recognition of beta 1,3-1,4-glucans by the family 11 carbohydrate-binding module from Clostridium thermocellum.
Febs J., 287, 2020
|
|
3KMV
 
 | | Crystal structure of CBM42A from Clostridium thermocellum | | Descriptor: | ACETATE ION, Alpha-L-arabinofuranosidase B, CALCIUM ION, ... | | Authors: | Santos-Silva, T, Alves, V.D, Prates, J.A.M, Fontes, C.M.G.A, Romao, M.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Family 42 carbohydrate-binding modules display multiple arabinoxylan-binding interfaces presenting different ligand affinities.
Biochim.Biophys.Acta, 1804, 2010
|
|
6R3M
 
 | | Family 11 Carbohydrate-Binding Module from Clostridium thermocellum in complex with beta-1,3-1,4-mixed-linked tetrasaccharide | | Descriptor: | ACETATE ION, CALCIUM ION, Endoglucanase H, ... | | Authors: | Ribeiro, D.O, Carvalho, A.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for the preferential recognition of beta 1,3-1,4-glucans by the family 11 carbohydrate-binding module from Clostridium thermocellum.
Febs J., 287, 2020
|
|
6SAA
 
 | | M-TRTX-Preg1a (Poecilotheria regalis) | | Descriptor: | U1-theraphotoxin-Pf3 | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-26 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
4BA6
 
 | | High Resolution structure of the C-terminal family 65 Carbohydrate Binding Module (CBM65B) of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | Endoglucanase cel5A, GLYCEROL | | Authors: | Venditto, I, Luis, A.S, Basle, A, Temple, M, Ferreira, L.M.A, Fontes, C.M.G.A, Gilbert, H.J, Najmudin, S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan.
J. Biol. Chem., 288, 2013
|
|
5FU2
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CALCIUM ION, CBM74-RFGH5, SODIUM ION, ... | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FU5
 
 | |
