4KR6
 
 | |
4KR7
 
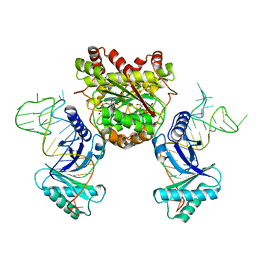 | | Crystal structure of a 4-thiouridine synthetase - RNA complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable tRNA sulfurtransferase, ... | | Authors: | Neumann, P, Ficner, R, Lakomek, K. | | Deposit date: | 2013-05-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.421 Å) | | Cite: | Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.
Nucleic Acids Res., 42, 2014
|
|
3G38
 
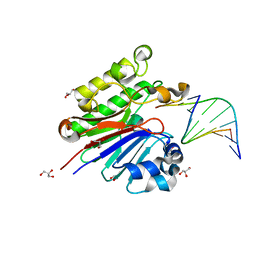 | | The catalytically inactive mutant Mth0212 (D151N) in complex with an 8 bp dsDNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*GP*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
4N6Q
 
 | | Crystal structure of VosA velvet domain | | Descriptor: | IODIDE ION, NITRATE ION, VosA | | Authors: | Ahmed, Y.L, Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Velvet Family of Fungal Regulators Contains a DNA-Binding Domain Structurally Similar to NF-kappa B.
Plos Biol., 11, 2013
|
|
4N3S
 
 | |
4N3G
 
 | |
4N3N
 
 | |
4NCN
 
 | |
4NCL
 
 | |
4NWI
 
 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 7-methylguanosine phosphate-specific 5'-nucleotidase, CHLORIDE ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
4NCF
 
 | |
4NOX
 
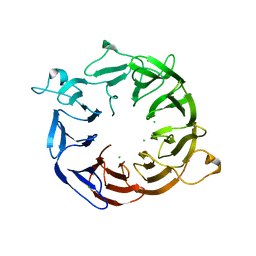 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
4NV0
 
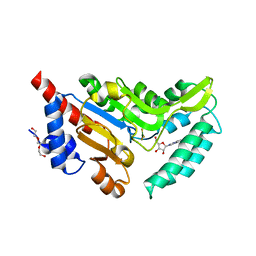 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to 7-methylguanosine | | Descriptor: | 7-METHYLGUANOSINE, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
3EGI
 
 | |
3FBX
 
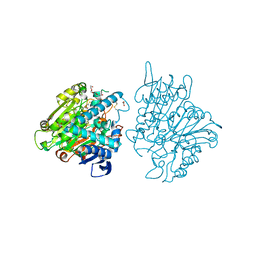 | | Crystal structure of the lysosomal 66.3 kDa protein from mouse solved by S-SAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Mueller, U, Ficner, R. | | Deposit date: | 2008-11-20 | | Release date: | 2009-03-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo sulfur SAD phasing of the lysosomal 66.3 kDa protein from mouse
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3G2D
 
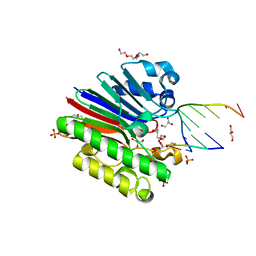 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G00
 
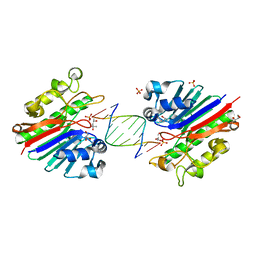 | | Mth0212 in complex with a 9bp blunt end dsDNA at 1.7 Angstrom | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*GP*TP*AP*TP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*UP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G0A
 
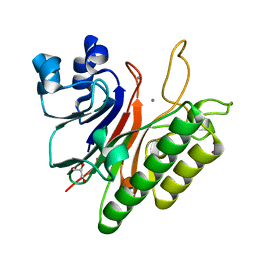 | | Mth0212 with two bound manganese ions | | Descriptor: | Exodeoxyribonuclease, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G2C
 
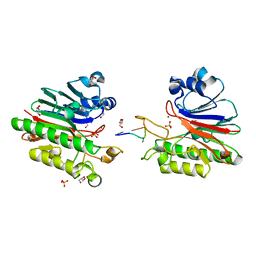 | | Mth0212 in complex with a short ssDNA (CGTA) | | Descriptor: | 5'-D(P*CP*GP*TP*A)-3', Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G4T
 
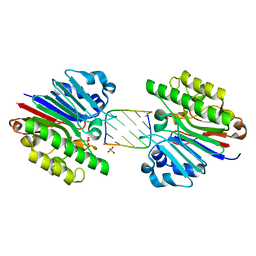 | | Mth0212 (WT) in complex with a 7bp dsDNA | | Descriptor: | 5'-D(*CP*G*TP*AP*CP*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-04 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G3C
 
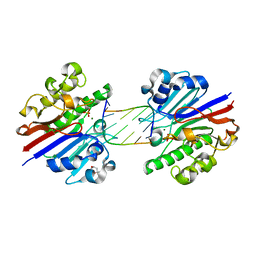 | | Mth0212 (WT) in complex with a 6bp dsDNA containing a single one nucleotide long 3'-overhang | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*CP*GP*TP*AP*(UPS)P*TP*AP*CP*G)-3', 5'-D(*CP*GP*TP*AP*CP*TP*AP*CP*G)-3', ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3FZI
 
 | |
3G0R
 
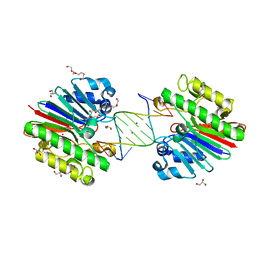 | | Complex of Mth0212 and an 8bp dsDNA with distorted ends | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*UP*GP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*CP*GP*CP*AP*GP*GP*GP*CP*G)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
3G1K
 
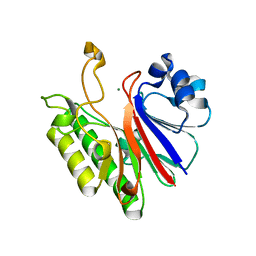 | |
3GDH
 
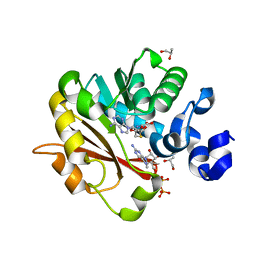 | | Methyltransferase domain of human Trimethylguanosine Synthase 1 (TGS1) bound to m7GTP and adenosyl-homocysteine (active form) | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, R-1,2-PROPANEDIOL, S-1,2-PROPANEDIOL, ... | | Authors: | Monecke, T, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1.
Nucleic Acids Res., 37, 2009
|
|
