8SCO
 
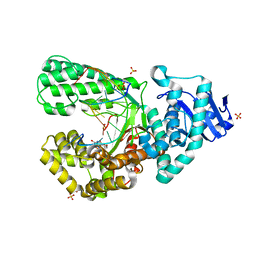 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 0h (Ground State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 3'-Amino DNA primer, CALCIUM ION, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCP
 
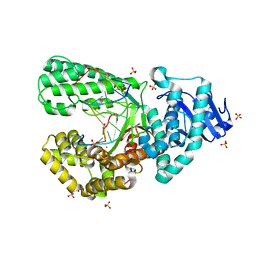 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 1h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCR
 
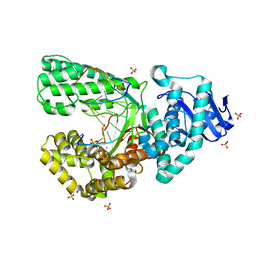 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 4h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCS
 
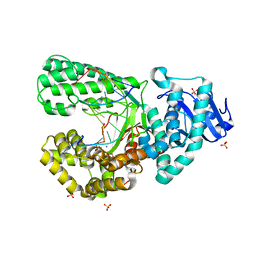 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCQ
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 2h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCT
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCU
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 48h (Product State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
7KUO
 
 | |
7KUN
 
 | |
7KUK
 
 | |
7KUM
 
 | |
7KUL
 
 | |
7KUP
 
 | |
7U8B
 
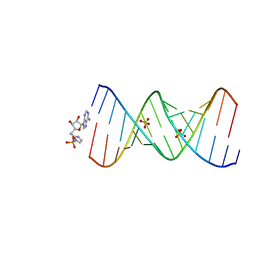 | | Product of 14mer primer with activated asymmetric GA dimer diastereomer 1 | | Descriptor: | 5'-O-[(S)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]adenosine, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3'), SULFATE ION | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Understanding the metal ion function in the RNA non-enzymatic extesion with phosphorothioates
To Be Published
|
|
4NOO
 
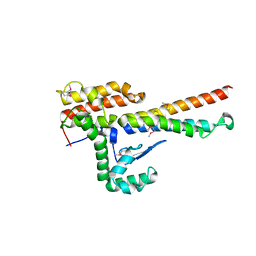 | | Molecular mechanism for self-protection against type VI secretion system in Vibrio cholerae | | Descriptor: | Putative uncharacterized protein, VgrG protein | | Authors: | Yang, X, Xu, M, Jiang, T, Fan, Z. | | Deposit date: | 2013-11-20 | | Release date: | 2014-04-09 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism for self-protection against the type VI secretion system in Vibrio cholerae.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
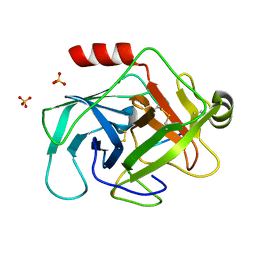 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
1AQK
 
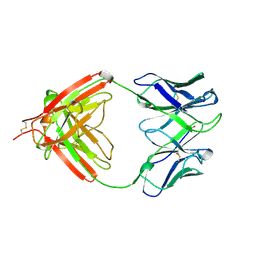 | |
1ETZ
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF AN ANTI-SWEETENER FAB, NC10.14, SHOWS THE EXTENT OF STRUCTURAL DIVERSITY IN ANTIGEN RECOGNITION BY IMMUNOGLOBULINS | | Descriptor: | FAB NC10.14 - HEAVY CHAIN, FAB NC10.14 - LIGHT CHAIN, N-(P-CYANOPHENYL)-N'-DIPHENYLMETHYL-GUANIDINE-ACETIC ACID | | Authors: | Guddat, L.W, Shan, L, Broomell, C, Ramsland, P.A, Fan, Z, Anchin, J.M, Linthicum, D.S, Edmundson, A.B. | | Deposit date: | 2000-04-13 | | Release date: | 2000-10-18 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The three-dimensional structure of a complex of a murine Fab (NC10. 14) with a potent sweetener (NC174): an illustration of structural diversity in antigen recognition by immunoglobulins.
J.Mol.Biol., 302, 2000
|
|
3U83
 
 | | Crystal structure of nectin-1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3U82
 
 | | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion | | Descriptor: | Envelope glycoprotein D, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.164 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
5EB3
 
 | | VB6-bound protein | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, SULFATE ION, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB0
 
 | | crystal form II of YfiB belonging to P41 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5EB2
 
 | | Trp-bound YfiR | | Descriptor: | SULFATE ION, TRYPTOPHAN, YfiR | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
