3NM0
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-(2,2-Difluoro-2-phenylethylamino)ethoxy) pyrrolidin-3-yl)methyl)-4-methyl-3,4,5,6-tetrahydropyridin-2-amine | | Descriptor: | (4S,6S)-6-{[(3R,4R)-4-{2-[(2,2-difluoro-2-phenylethyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methyl-3,4,5,6-tetrahydropyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLU
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2,2-difluoro-2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLD
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6-{{(3'S,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Ji, H, Delker, S.L, Li, H, Martasek, P, Roman, L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3NLT
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-{(3'S,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}- N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({(3S,4S)-4-[2-({2,2-difluoro-2-[(2R)-piperidin-2-yl]ethyl}amino)ethoxy]pyrrolidin-3-yl}methyl)-4-methylpyridin-2-amine, CACODYLIC ACID, ... | | Authors: | Xue, F, Li, H, Fang, J, Delker, S.L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLP
 
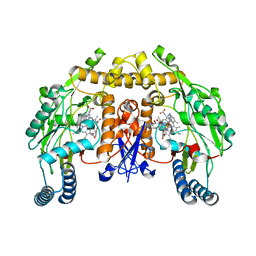 | | Structure of neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with 6-{{(3'S,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3NLF
 
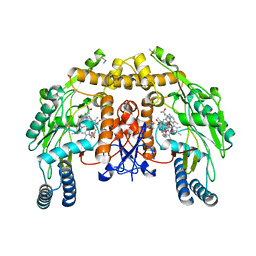 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6-{{(3'R,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4R)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Ji, H, Delker, S.L, Li, H, Martasek, P, Roman, L, Poulos, T.L, Silverman, R.B. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3NLR
 
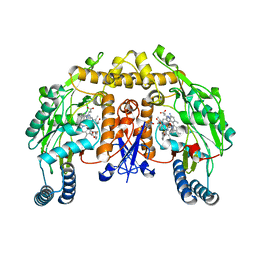 | | Structure of neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with 6-{{(3'R,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4R)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3NLW
 
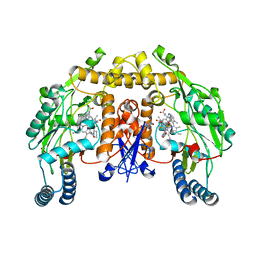 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-(2,2-Difluoro-2-(piperidin-2-yl)ethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({(3S,4S)-4-[2-({2,2-difluoro-2-[(2R)-piperidin-2-yl]ethyl}amino)ethoxy]pyrrolidin-3-yl}methyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Potent, highly selective, and orally bioavailable gem-difluorinated monocationic inhibitors of neuronal nitric oxide synthase.
J.Am.Chem.Soc., 132, 2010
|
|
3NLO
 
 | | Structure of neuronal nitric oxide synthase R349A mutant heme domain in complex with 6-{{(3'S,4'R)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4S)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
7JFZ
 
 | |
7K72
 
 | |
6MJK
 
 | |
5IFZ
 
 | |
6CY1
 
 | |
6CY5
 
 | |
6E54
 
 | |
6B8V
 
 | |
6C9C
 
 | |
6C6B
 
 | |
5UZX
 
 | |
5VA8
 
 | |
5UMH
 
 | |
5VWM
 
 | |
5VJY
 
 | |
5VPU
 
 | |
